5G4Y
 
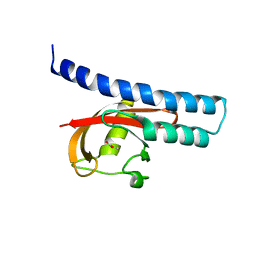 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
5G26
 
 | | Unveiling the Mechanism Behind the in-meso Crystallization of Membrane Proteins | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, INTIMIN | | Authors: | Zabara, A, Newman, J, Peat, T.S. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Nanoscience Behind the Art of in-Meso Crystallization of Membrane Proteins.
Nanoscale, 9, 2017
|
|
4AHT
 
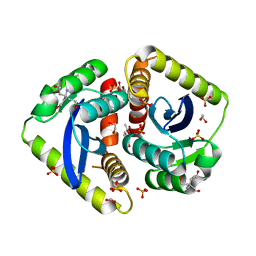 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHU
 
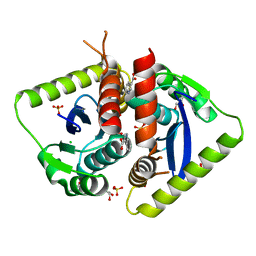 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOLE-3-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4ADD
 
 | |
4ADC
 
 | |
4AHV
 
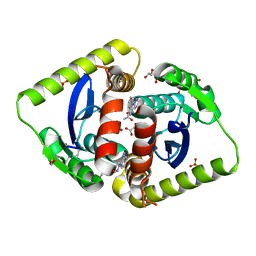 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(1H-pyrazol-1-yl)phenyl]methanamine, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHR
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 3-(1,3-benzodioxol-5-yl)propanoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4ADE
 
 | |
4ADB
 
 | |
4AH9
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHS
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZOFURAN-7-CARBOXYLIC ACID, ACETATE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4BJO
 
 | | Nitrate in the active site of PTP1b is a putative mimetic of the transition state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kenny, P.W, Newman, J, Peat, T.S. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Nitrate in the Active Site of Protein Tyrosine Phosphatase 1B is a Putative Mimetic of the Transition State.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CRJ
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP and an inhibitor | | Descriptor: | 2-amino-8-{[2-(4-methoxyphenyl)-2-oxoethyl]sulfanyl}-1,9-dihydro-6H-purin-6-one, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE (HPPK), DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
4CQC
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CWB
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP and an inhibitor | | Descriptor: | 2-amino-8-[2-oxo-2-(4-phenylphenyl)ethyl]sulfanyl-1,9-dihydropurin-6-one, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4CYY
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYG
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | (2R)-2,4-dihydroxy-N-[(3S)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNV
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CYF
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYU
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-15 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4D70
 
 | | Structural, biophysical and biochemical analyses of a Clostridium perfringens Sortase D5 transpeptidase | | Descriptor: | SORTASE FAMILY PROTEIN | | Authors: | Suryadinata, R, Seabrook, S, Adams, T.E, Nuttall, S.D, Peat, T.S. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Biochemical Analyses of a Clostridium Perfringens Sortase D Transpeptidase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CNW
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CALCIUM ION, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CQ0
 
 | | Cyclic secondary sulfonamides: unusually good inhibitors of cancer- related carbonic anhydrase enzymes | | Descriptor: | 6-amino-1,2-benzothiazol-3(2H)-one 1,1-dioxide, CARBONIC ANHYDRASE 2, FORMIC ACID, ... | | Authors: | Moeker, J, Peat, T.S, Bornaghi, L.F, Vullo, D, Supuran, C.T, Poulsen, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Cyclic Secondary Sulfonamides: Unusually Good Inhibitors of Cancer-Related Carbonic Anhydrase Enzymes.
J.Med.Chem., 57, 2014
|
|
