4NFJ
 
 | | Crystal structure of human FPPS in complex with magnesium, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Park, J, De Schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4NFI
 
 | | Crystal structure of human FPPS in complex with magnesium and JDS05120 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [({5-[4-(cyclopropyloxy)phenyl]pyridin-3-yl}amino)methanediyl]bis(phosphonic acid) | | Authors: | Park, J, De Schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4NFK
 
 | | Crystal structure of human FPPS in complex with nickel, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Park, J, De schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4OF1
 
 | |
4QXS
 
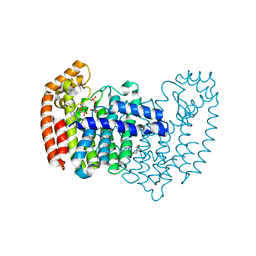 | | Crystal structure of human FPPS in complex with WC01088 | | Descriptor: | (2-{2-[(2S)-3-methylbutan-2-yl]-5-phenyl-1H-indol-3-yl}ethane-1,1-diyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Park, J, Zielinski, M, Weiling, C, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the molecular and structural elements of ligands binding to the active site versus an allosteric pocket of the human farnesyl pyrophosphate synthase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4QY5
 
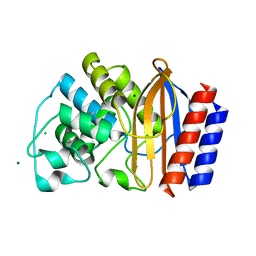 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
4ZEV
 
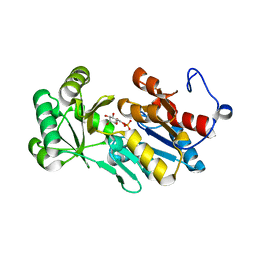 | | Crystal structure of PfHAD1 in complex with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZEW
 
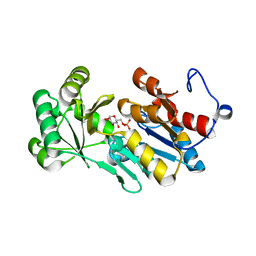 | | Crystal structure of PfHAD1 in complex with glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZEX
 
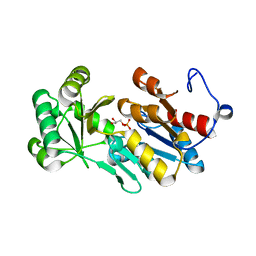 | | Crystal structure of PfHAD1 in complex with glyceraldehyde-3-phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6A0H
 
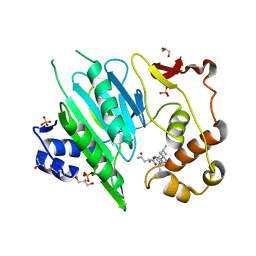 | |
6A0F
 
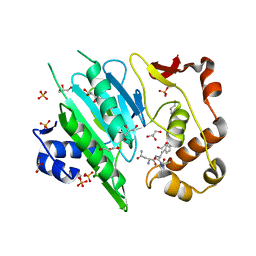 | |
6A0I
 
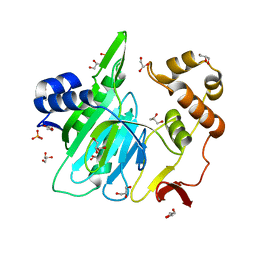 | |
6A0E
 
 | |
6K17
 
 | | Crystal structure of EXD2 exonuclease domain | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, SODIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1E
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1B
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6KY2
 
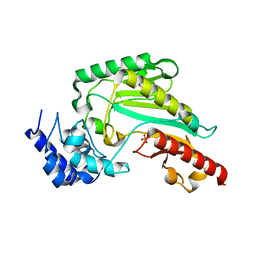 | | Crystal Structure of Arginine Kinase wild type from Daphnia magna | | Descriptor: | Arginine kinase, PHOSPHATE ION | | Authors: | Park, J.H, Rao, Z, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6N7Y
 
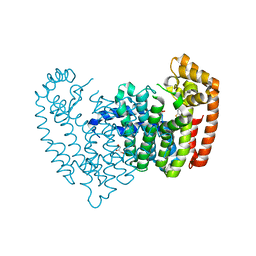 | |
6N83
 
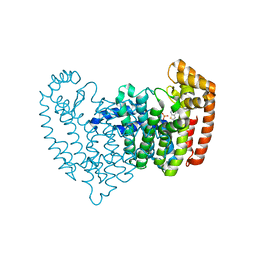 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, PHOSPHATE ION, ... | | Authors: | Park, J, Schilling, M.A, Berghuis, A.M. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
6N7Z
 
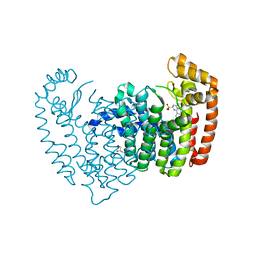 | |
6OAH
 
 | |
4LFV
 
 | | Crystal structure of human FPPS in complex with YS0470 and two molecules of inorganic phosphate | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human farnesyl pyrophosphate synthase in complex with an aminopyridine bisphosphonate and two molecules of inorganic phosphate.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
6OAG
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
5XYK
 
 | | Structure of Transferase | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J, Cho, H.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Transferase
To Be Published
|
|
