1GE6
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE5
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE7
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1WVV
 
 | |
1CYD
 
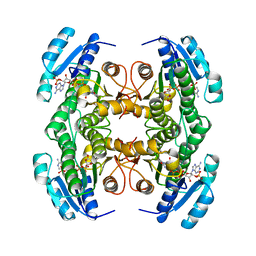 | | CARBONYL REDUCTASE COMPLEXED WITH NADPH AND 2-PROPANOL | | Descriptor: | CARBONYL REDUCTASE, ISOPROPYL ALCOHOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family.
Structure, 4, 1996
|
|
4Y04
 
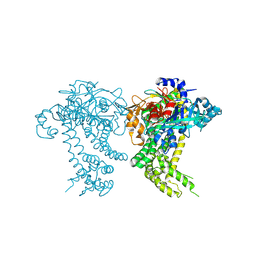 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Space) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Inaka, K, Tanaka, H, Yamada, M, Ohta, K, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4XZY
 
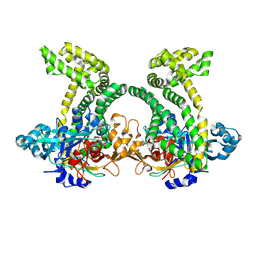 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y02
 
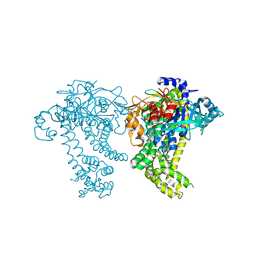 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Ground) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y01
 
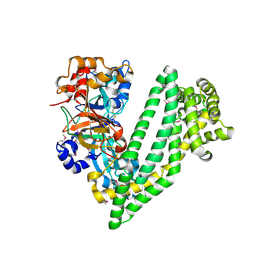 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
1KW6
 
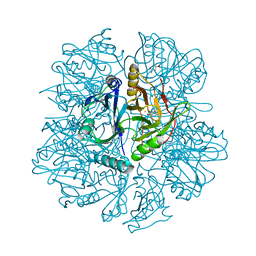 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 1.45 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW3
 
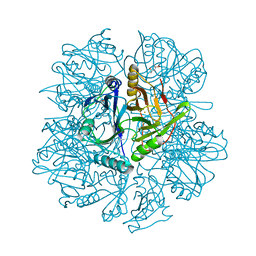 | | Crystal structure of 2,3-dihydroxybiphenyal dioxygenase (BphC) at 1.45 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-Dihydroxybiphenyl dioxygenase, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KWC
 
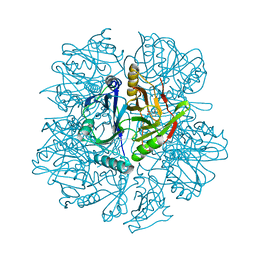 | | The His145Ala mutant of 2,3-dihydroxybiphenyl dioxygenase in complex with 2,3-dihydroxybiphenyl | | Descriptor: | 2,3-dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW9
 
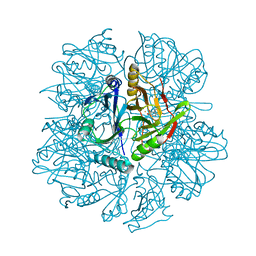 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 2.0A resolution | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KWB
 
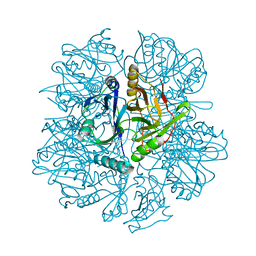 | | Crystal structure of the His145Ala mutant of 2,3-dihydroxybipheny dioxygenase (BphC) | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW8
 
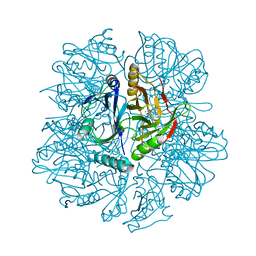 | | Crystal structure of BphC-2,3-dihydroxybiphenyl-NO complex | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
2DKV
 
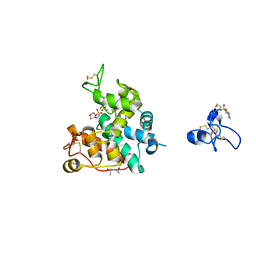 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
5XEM
 
 | |
7DKD
 
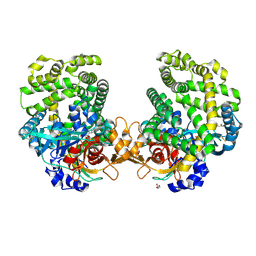 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
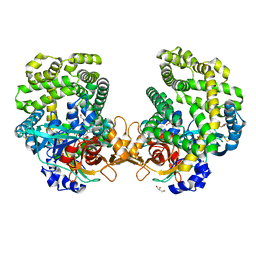 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
5XEO
 
 | |
5XEN
 
 | |
6IOY
 
 | | Crystal structure of Porphyromonas gingivalis acetate kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetate kinase, SULFATE ION | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2018-10-31 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production inPorphyromonas gingivalis.
J Oral Microbiol, 11, 2019
|
|
