11AS
 
 | |
12AS
 
 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
2D1Q
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1R
 
 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | Descriptor: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1S
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1T
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2AE2
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADP+ AND PSEUDOTROPINE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (TROPINONE REDUCTASE-II), PSEUDOTROPINE | | Authors: | Yamashita, A, Kato, H, Wakatsuki, S, Tomizaki, T, Nakatsu, T, Nakajima, K, Hashimoto, T, Yamada, Y, Oda, J. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
Biochemistry, 38, 1999
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
7FC9
 
 | | Crystal structure of CmABCB1 in lipidic mesophase revealed by LCP-SFX | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pan, D, Oyama, R, Sato, T, Nakane, T, Mizunuma, R, Matsuoka, K, Joti, Y, Tono, K, Nango, E, Iwata, S, Nakatsu, T, Kato, H. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.
Iucrj, 9, 2022
|
|
2AE1
 
 | | TROPINONE REDUCTASE-II | | Descriptor: | TROPINONE REDUCTASE-II | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6A6M
 
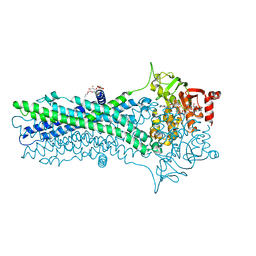 | | Crystal structure of an outward-open nucleotide-bound state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
6A6N
 
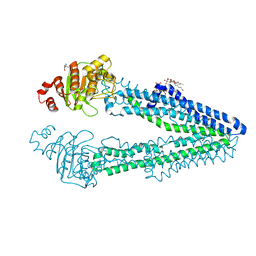 | | Crystal structure of an inward-open apo state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
3ED1
 
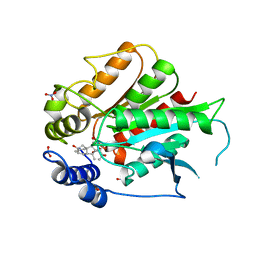 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
1IPE
 
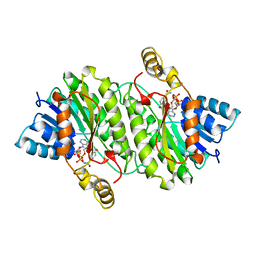 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1IPF
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH AND TROPINONE | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
3EBL
 
 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
7DQV
 
 | | Crystal structure of a CmABCB1 mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Matsuoka, K, Nakatsu, T, Kato, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the CmABCB1 G132V mutant, which favors the outward-facing state, reveals the mechanism of the pivotal joint between TM1 and TM3.
Protein Sci., 30, 2021
|
|
5GX3
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 6.9 MGy (6th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX5
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 26 MGy (23rd measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX1
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 1.1 MGy (1st measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX2
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 3.4 MGy (3rd measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX4
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 14 MGy (12th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
1KCD
 
 | | Endopolygalacturonase I from Stereum purpureum complexed with two galacturonate at 1.15 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1KCC
 
 | | Endopolygalacturonase I from Stereum purpureum complexed with a galacturonate at 1.00 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1K5C
 
 | | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-10-10 | | Release date: | 2002-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
