7K2O
 
 | | Kelch domain of human KEAP1 bound to Nrf2-based cyclic peptide, c[GABA-DPETGE] | | Descriptor: | (ABU)DPETGE, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2H
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GDPETGE] | | Descriptor: | GLY-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2M
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GEPETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GEPETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2J
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GDPEAGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GDPEAGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K28
 
 | | Kelch domain of human KEAP1 bound to Nrf2 peptide, ADEETGEFL | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 peptide,ADEETGEFL | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2A
 
 | | Kelch domain of human KEAP1 bound to Nrf2 peptide, LDEETGEFA | | Descriptor: | ACE-LEU-ASP-GLU-GLU-THR-GLY-GLU-PHE-ALA-NH2, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2K
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[BAL-DEETGE] | | Descriptor: | BAL-ASP-GLU-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2E
 
 | | Kelch domain of human KEAP1 bound to Nrf2-based cyclic peptide, c[GDEETGE] | | Descriptor: | GLY-ASP-GLU-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2G
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GDEEAGE] | | Descriptor: | GLY-ASP-GLU-GLU-ALA-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2S
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[DhA-GDPETGE] | | Descriptor: | B3A-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2P
 
 | | Kelch domain of human KEAP1 bound to Nrf2-based cyclic peptide, c[AVA-DPETGE] | | Descriptor: | (DAV)DPETGE, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2L
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[BAL-NPETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[BAL-NPETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2F
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GAEETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GAEETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2I
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GAPETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GAPETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2D
 
 | | Kelch domain of human KEAP1 bound to Nrf2 linear peptide, Ac-GDEETGE-NH2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 linear peptide, Ace-GDEETGE-NH2 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2Q
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[Ahx-DPETGE] | | Descriptor: | ACA-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
2J8H
 
 | | Structure of the immunoglobulin tandem repeat A168-A169 of titin | | Descriptor: | GLYCEROL, TITIN | | Authors: | Mueller, S, Lange, S, Kursula, I, Gautel, M, Wilmanns, M. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-Band of the Elastic Muscle Protein Titin
J.Mol.Biol., 371, 2007
|
|
2J8O
 
 | | Structure of the immunoglobulin tandem repeat of titin A168-A169 | | Descriptor: | GLYCEROL, TITIN | | Authors: | Mueller, S, Lange, S, Kursula, I, Gautel, M, Wilmanns, M. | | Deposit date: | 2006-10-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-Band of the Elastic Muscle Protein Titin
J.Mol.Biol., 371, 2007
|
|
2BK8
 
 | |
2BKF
 
 | |
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFQ
 
 | | Crystal Structure of Exocytosis-Sensitive Phosphoprotein, pp63/parafusin (Phosphoglucomutse) from Paramecium. OPEN FORM | | Descriptor: | CALCIUM ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-22 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
2WY3
 
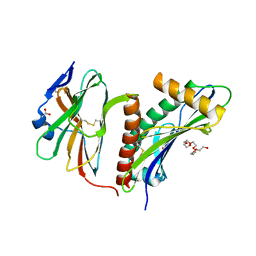 | | Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Mueller, S, Zocher, G, Steinle, A, Stehle, T. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Hcmv Ul16-Micb Complex Elucidates Select Binding of a Viral Immunoevasin to Diverse Nkg2D Ligands.
Plos Pathog., 6, 2010
|
|
1DGI
 
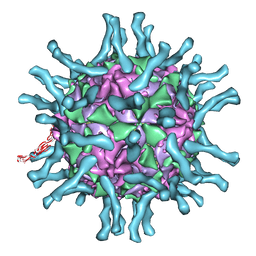 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1NN8
 
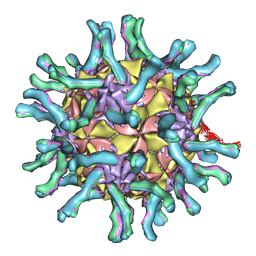 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
