8T2H
 
 | | DYRK1A complex with DYR530 | | Descriptor: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | Authors: | Montfort, W.R, Basantes, L.E. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of DYR684, a Potent, Selective, Metabolically Stable, DYRK1A/B PROTAC utilizing a Novel Cereblon Molecular Glue
To Be Published
|
|
3FLL
 
 | | Crystal structure of E55Q mutant of nitrophorin 4 | | Descriptor: | AMMONIA, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Montfort, W.R, Weichsel, A. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of mutation of carboxyl side-chain amino acids near the heme on the midpoint potentials and ligand binding constants of nitrophorin 2 and its NO, histamine, and imidazole complexes.
J.Am.Chem.Soc., 131, 2009
|
|
2FTN
 
 | | E. coli thymidylate synthase Y94F mutant | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Montfort, W.R, Roberts, S.A. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Y94F mutant of Escherichia coli thymidylate synthase.
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2FTQ
 
 | | E. coli thymidylate synthase at 1.8 A resolution | | Descriptor: | PHOSPHATE ION, SULFATE ION, Thymidylate synthase | | Authors: | Montfort, W.R, Roberts, S.A. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Subtle Conformational Differences between Escherichia coli Thymidylate Synthase and Specific Mutants of this Enzyme
To be Published
|
|
2TSC
 
 | | STRUCTURE, MULTIPLE SITE BINDING, AND SEGMENTAL ACCOMODATION IN THYMIDYLATE SYNTHASE ON BINDING D/UMP AND AN ANTI-FOLATE | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Montfort, W.R, Stroud, R.M. | | Deposit date: | 1991-07-03 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate.
Biochemistry, 29, 1990
|
|
3QQX
 
 | | Reduced Native Intermediate of the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CueO E506D Mutant: Crystal Structure of Reduced Native Intermediate, Kinetics, and Impairment of Product Release
To be Published
|
|
3NSD
 
 | | Silver bound to the multicopper oxidase CueO (untagged) | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, OXYGEN ATOM, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3OD3
 
 | | CueO at 1.1 A resolution including residues in previously disordered region | | Descriptor: | 1,2-ETHANEDIOL, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2010-08-10 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3PAV
 
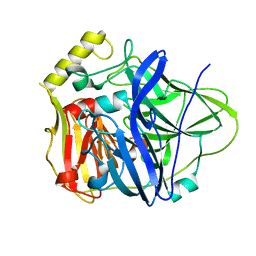 | |
4NP1
 
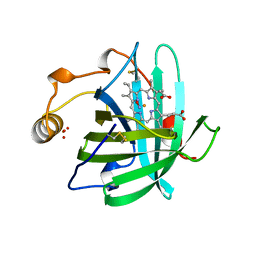 | | NITROPHORIN 1 COMPLEX WITH NITRIC OXIDE | | Descriptor: | NITRIC OXIDE, NITROPHORIN 1, PHOSPHATE ION, ... | | Authors: | Weichsel, A, Andersen, J.F, Walker, F.A, Montfort, W.R. | | Deposit date: | 1998-06-26 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitric Oxide Binding to the Ferri-and Ferroheme States of Nitrophorin 1, a Reversible No-Binding Heme Protein from the Saliva of the Blood-Sucking Insect, Rhodnius Prolixus
To be Published
|
|
3C76
 
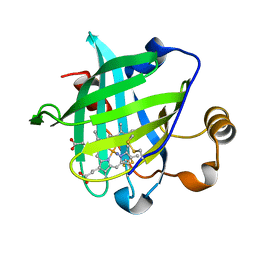 | |
3C78
 
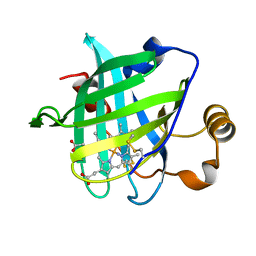 | |
6WQE
 
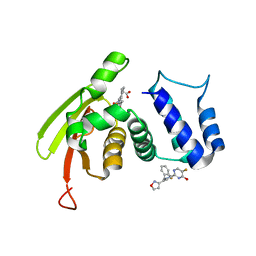 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
2FTO
 
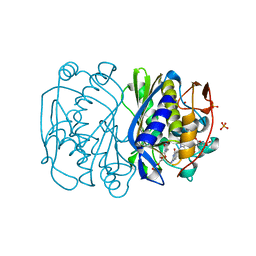 | | Y94F mutant of thymidylate synthase bound to thymidine-5'-phosphate and 10-propargyl-5,8-dideazafolid acid | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, PHOSPHATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Roberts, S.A, Montfort, W.R. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Y94F mutant of Escherichia coli thymidylate synthase.
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
4GRJ
 
 | |
4OO5
 
 | |
4OO4
 
 | |
3UKO
 
 | |
6DXI
 
 | |
3FGC
 
 | | Crystal Structure of the Bacterial Luciferase:Flavin Complex Reveals the Basis of Intersubunit Communication | | Descriptor: | Alkanal monooxygenase alpha chain, Alkanal monooxygenase beta chain, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Campbell, Z.T, Weichsel, A, Montfort, W.R, Baldwin, T.O. | | Deposit date: | 2008-12-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit.
Biochemistry, 48, 2009
|
|
6MX5
 
 | |
6OCV
 
 | |
4GNW
 
 | |
1FFL
 
 | |
3M9K
 
 | | Crystal structure of human thioredoxin C69/73S double-mutant, oxidized form | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, SULFATE ION, Thioredoxin | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2010-03-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human thioredoxin revealing an unraveled helix and exposed S-nitrosation site.
Protein Sci., 19, 2010
|
|
