1EGF
 
 | |
2SPZ
 
 | | STAPHYLOCOCCAL PROTEIN A, Z-DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Montelione, G.T, Tashiro, M, Tejero, R, Lyons, B.A. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the Z domain of staphylococcal protein A.
J.Mol.Biol., 272, 1997
|
|
1NS1
 
 | |
3EGF
 
 | |
7VYQ
 
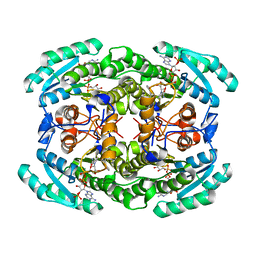 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
1MV4
 
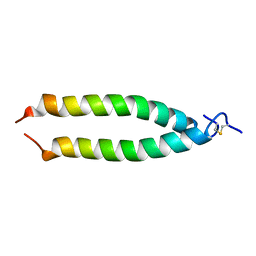 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | Descriptor: | Tropomyosin 1 alpha chain | | Authors: | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | Deposit date: | 2002-09-24 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
4ZEQ
 
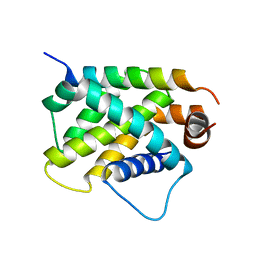 | | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide, Northeast Structural Genomics Consortium Target HX9247 | | Descriptor: | BH3-interacting domain death agonist, Bcl-2-related protein A1 | | Authors: | Guan, R, Xiao, R, Mao, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide
To Be Published
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
6E4J
 
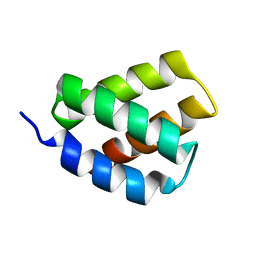 | | Solution NMR Structure of Protein PF2048.1 | | Descriptor: | Uncharacterized protein PF2048.1 | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
7K3S
 
 | |
5GAJ
 
 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | Descriptor: | DE NOVO DESIGNED PROTEIN OR258 | | Authors: | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
2FVT
 
 | | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris: Northeast Structural Genomics Target RpR43 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Ho, C.K, Cunningham, K, Ma, L.C, Conover, K, Xiao, R, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris
To be Published
|
|
2GBS
 
 | | NMR structure of Rpa0253 from Rhodopseudomonas palustris. Northeast structural genomics consortium target RpR3 | | Descriptor: | Hypothetical protein Rpa0253 | | Authors: | Ramelot, T.A, Cort, J.R, Conover, K, Chen, Y, Ma, L.C, Ciano, M, Xiao, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-11 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
8CWA
 
 | |
8CTO
 
 | |
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
6E4H
 
 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
4RZP
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR366. | | Descriptor: | Engineered Protein OR366 | | Authors: | Vorobiev, S, Parmeggiani, F, Dimaio, F, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of Engineered Protein OR366
To be Published
|
|
4RMM
 
 | | Crystal Structure of the Q7NVP2_CHRVO protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR191 | | Descriptor: | Putative uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-21 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Q7NVP2_CHRVO protein from Chromobacterium violaceum.
To be Published
|
|
4RV1
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR497. | | Descriptor: | ACETATE ION, Engineered Protein OR497 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Crystal Structure of Engineered Protein OR497.
To be Published
|
|
4RL6
 
 | | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR105 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Vorobiev, S, Neely, H.M, Odukwe, C.D, Seetharaman, J, Mao, L, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae.
To be Published
|
|
7M5T
 
 | |
1AIL
 
 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | Descriptor: | NONSTRUCTURAL PROTEIN NS1 | | Authors: | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | Deposit date: | 1997-04-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
5DIL
 
 | | Crystal structure of the effector domain of the NS1 protein from influenza virus B | | Descriptor: | IODIDE ION, Non-structural protein 1 | | Authors: | Guan, R, Hamilton, K, Ma, L, Montelione, G.T. | | Deposit date: | 2015-09-01 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Second RNA-Binding Site in the NS1 Protein of Influenza B Virus.
Structure, 24, 2016
|
|
