8I6V
 
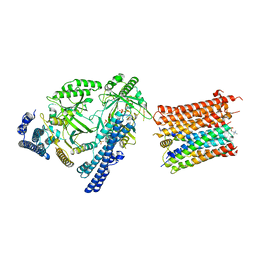 | | Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mayer, A, Wu, S, Ye, S. | | Deposit date: | 2023-01-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit.
Embo J., 42, 2023
|
|
8AFW
 
 | | Tube assembly of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFQ
 
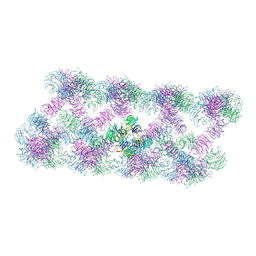 | | Tube assembly of Atg18-PR72AA | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFY
 
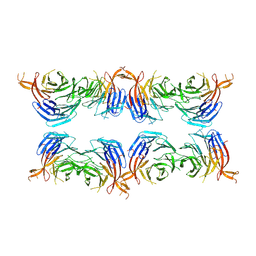 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFX
 
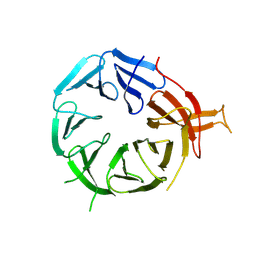 | | Single particle structure of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures of Atg18 oligomers reveal a tilted structural scaffold for Atg2 at the isolation membrane
To Be Published
|
|
5CE7
 
 | | Structure of a non-canonical CID of Ctk3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CTD kinase subunit gamma | | Authors: | Muehlbacher, W, Mayer, A, Sun, M, Remmert, M, Cheung, A.C, Niesser, J, Soeding, J, Cramer, P. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ctk3, a subunit of the RNA polymerase II CTD kinase complex, reveals a noncanonical CTD-interacting domain fold.
Proteins, 83, 2015
|
|
3GXW
 
 | | Structure of the SH2 domain of the Candida glabrata transcription elongation factor Spt6, crystal form A | | Descriptor: | SODIUM ION, SUCCINIC ACID, Transcription elongation factor SPT6 | | Authors: | Dengl, S, Mayer, A, Sun, M, Cramer, P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and in vivo requirement of the yeast Spt6 SH2 domain
J.Mol.Biol., 389, 2009
|
|
3TJ1
 
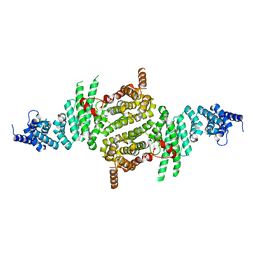 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
3GXX
 
 | | Structure of the SH2 domain of the Candida glabrata transcription elongation factor Spt6, crystal form B | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Dengl, S, Mayer, A, Sun, M, Cramer, P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and in vivo requirement of the yeast Spt6 SH2 domain
J.Mol.Biol., 389, 2009
|
|
3PJP
 
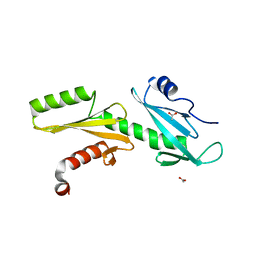 | | A Tandem SH2 Domain in Transcription Elongation Factor Spt6 Binds the Phosphorylated RNA Polymerase II C-terminal Repeat Domain(CTD) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Transcription elongation factor SPT6 | | Authors: | Sun, M, Lariviere, L, Dengl, S, Mayer, A, Cramer, P. | | Deposit date: | 2010-11-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD).
J.Biol.Chem., 285, 2010
|
|
