7Q2Z
 
 | |
5F0O
 
 | | Cohesin subunit Pds5 in complex with Scc1 | | Descriptor: | KLTH0G16610p, cohesin subunit Pds5, KLTH0D07062p,KLTH0D07062p,KLTH0D07062p,cohesin subunit Pds5, ... | | Authors: | Lee, B.-G, Jansma, M, Nasmyth, K, Lowe, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1.
Cell Rep, 14, 2016
|
|
7QUD
 
 | | D. melanogaster alpha/beta tubulin heterodimer in the GTP form | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tubulin alpha-1 chain, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUC
 
 | | D. melanogaster alpha/beta tubulin heterodimer in the GDP form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUQ
 
 | | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii) | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin beta, Tubulin domain-containing protein | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUP
 
 | | D. melanogaster 13-protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
8AFL
 
 | |
8AIX
 
 | |
8AFM
 
 | |
8AIA
 
 | |
8AFE
 
 | |
7NYY
 
 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
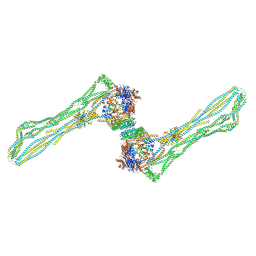 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYX
 
 | |
7NZ2
 
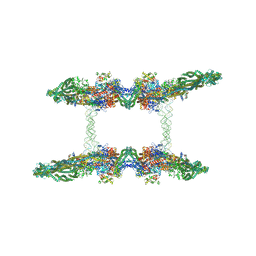 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYW
 
 | | Cryo-EM structure of the MukBEF-MatP-DNA head module | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYZ
 
 | |
7NZ0
 
 | |
7NZ3
 
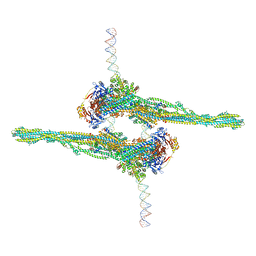 | | Cryo-EM structure of apposed MukBEF-MatP monomers on DNA | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
6RIQ
 
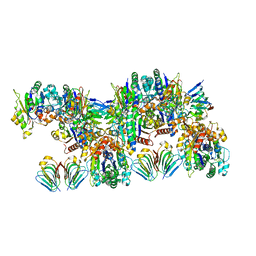 | | MinCD filament from Pseudomonas aeruginosa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MinC, ... | | Authors: | Szewczak-Harris, A, Wagstaff, J, Lowe, J. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the MinCD copolymeric filament from Pseudomonas aeruginosa at 3.1 angstrom resolution.
Febs Lett., 593, 2019
|
|
5AI7
 
 | | ParM doublet model | | Descriptor: | PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles
Nature, 523, 2015
|
|
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1HF2
 
 | |
5AEY
 
 | | actin-like ParM protein bound to AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles.
Nature, 523, 2015
|
|
6SCJ
 
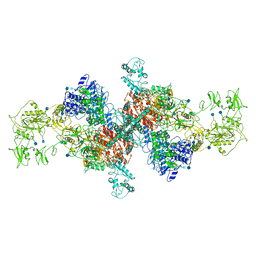 | | The structure of human thyroglobulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Coscia, F, Turk, D, Lowe, J. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structure of human thyroglobulin.
Nature, 578, 2020
|
|
