3WQM
 
 | | Crystal structure of Rv3378c with inhibitor BPH-629 | | Descriptor: | Diterpene synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
7XQS
 
 | | The structure of FLA-K*00701/KP-CoV-9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQT
 
 | | The structure of FLA-K*00701/KP-FECV-11 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQU
 
 | | The structure of FLA-E*00301/EM-FECV-10 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide from Nucleoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRE
 
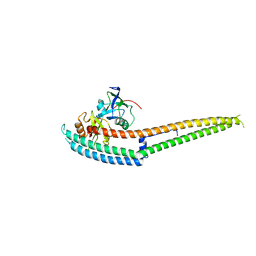 | | Complex structure of INAD PDZ45 and NORPA CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, Inactivation-no-after-potential D protein | | Authors: | Ye, F, Li, J, Deng, X, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
7CMA
 
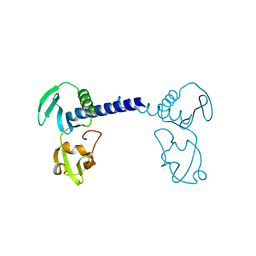 | | Structure of A151R from African swine fever virus Georgia | | Descriptor: | A151R, ZINC ION | | Authors: | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | Deposit date: | 2020-07-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
5UVJ
 
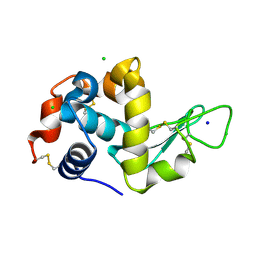 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
6IEX
 
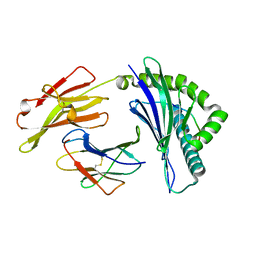 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
2H1Y
 
 | |
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5CUY
 
 | | Crystal structure of Trypanosoma brucei Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, CITRIC ACID, MAGNESIUM ION | | Authors: | Yang, Y.Y, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
5CUV
 
 | | Crystal structure of Trypanosoma cruzi Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, D-MALATE, MAGNESIUM ION | | Authors: | Ko, T.P, Yang, Y.Y, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
5XJX
 
 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
7X3A
 
 | | NMR solution structure of the 1:1 complex of a pyridostatin (PDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X2Z
 
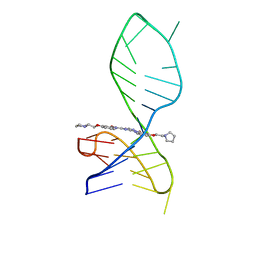 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
8J70
 
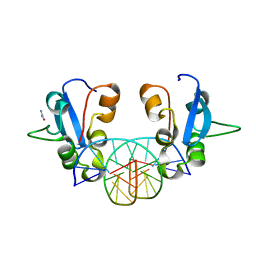 | | Native SAND domain of protein SP140 with DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*GP*CP*CP*GP*CP*CP*CP*T)-3'), Nuclear body protein SP140, selenourea | | Authors: | Li, H.T, Liu, W.Q. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for Speckled protein SP140 bivalent recognition of histone H3 and DNA.
To Be Published
|
|
8J71
 
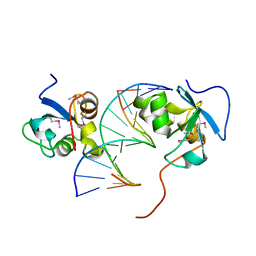 | |
6JLE
 
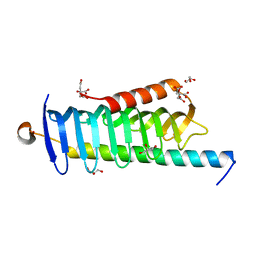 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
8GZD
 
 | |
5ZUM
 
 | | Structure of dipeptidyl-peptidase III from Corallococcus sp. strain EGB | | Descriptor: | ZINC ION, dipeptidyl-peptidase III | | Authors: | Zhang, H, Duan, Y.J, Li, Z.K, Liu, W.D, Huang, Y, Cui, Z.L. | | Deposit date: | 2018-05-08 | | Release date: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of dipeptidyl peptidase III from Corallococcus sp. strain EGB
To Be Published
|
|
6AIN
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIO
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6IRC
 
 | | C-terminal domain of Drosophila phospholipase b NORPA, methylated | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
7CWQ
 
 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
