5N9D
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with GGGTTAGGGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9A
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*TP*TP*AP*GP*GP*GP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.036 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8S
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with polyT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N96
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*AP*GP*GP*GP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8R
 
 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N90
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
1REO
 
 | | L-amino acid oxidase from Agkistrodon halys pallas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AHPLAAO, CITRIC ACID, ... | | Authors: | Zhang, H, Teng, M, Niu, L, Wang, Y, Wang, Y, Liu, Q, Huang, Q, Hao, Q, Dong, Y, Liu, P. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Purification, partial characterization, crystallization and structural determination of AHP-LAAO, a novel L-amino-acid oxidase with cell apoptosis-inducing activity from Agkistrodon halys pallas venom.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5N8Z
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCCTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.477 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N8U
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N9E
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TGGGGATTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*GP*GP*GP*GP*AP*TP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N9F
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A | | Descriptor: | CG9323, isoform A, CHLORIDE ION, ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.969 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
7SEU
 
 | | Crystal structure of E coli contaminant protein YncE co-purified with a plant protein | | Descriptor: | PQQ-dependent catabolism-associated beta-propeller protein | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
4WD7
 
 | |
4U9A
 
 | |
1Z6L
 
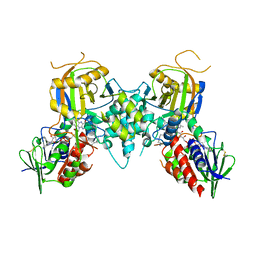 | |
4RYO
 
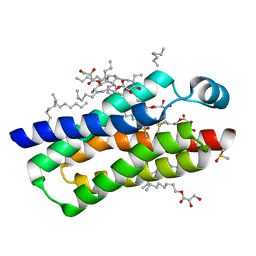 | | Crystal structure of BcTSPO type II high resolution monomer | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4WD8
 
 | |
1V6P
 
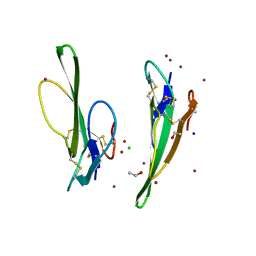 | | Crystal structure of Cobrotoxin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Cobrotoxin, ... | | Authors: | Lou, X, Tu, X, Wang, J, Teng, M, Niu, L, Liu, Q, Huang, Q, Hao, Q. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
1XPQ
 
 | | Crystal structure of fms1, a polyamine oxidase from yeast | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, SPERMINE | | Authors: | Huang, Q, Liu, Q, Hao, Q. | | Deposit date: | 2004-10-09 | | Release date: | 2005-04-26 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J.Mol.Biol., 348, 2005
|
|
1YY5
 
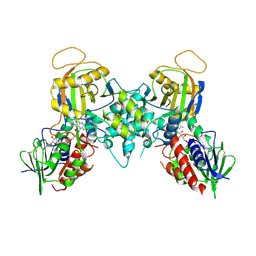 | |
6W8S
 
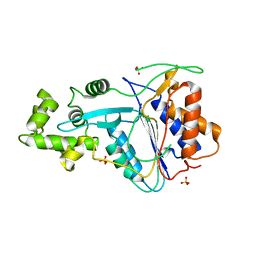 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8T
 
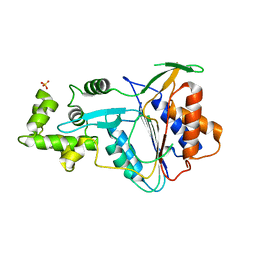 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8R
 
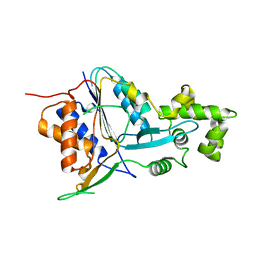 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
8X6F
 
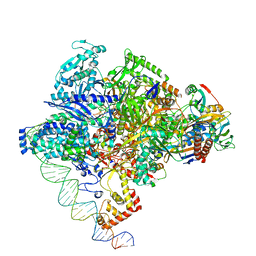 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase
Nat Commun, 2024
|
|
8X6G
 
 | | Cryo-EM structure of Staphylococcus aureus sigB-dependent RNAP-promoter open complex | | Descriptor: | DNA (70-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase
Nat Commun, 2024
|
|
