7N6Q
 
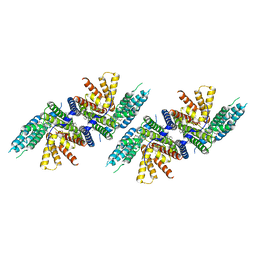 | | Structure of PPPA bound human ACAT2 | | Descriptor: | (3S,4R,4aR,6S,6aS,12R,12aS,12bS)-4-[(acetyloxy)methyl]-12-hydroxy-4,6a,12b-trimethyl-11-oxo-9-(pyridin-3-yl)-1,3,4,4a,5,6,6a,12,12a,12b-decahydro-2H,11H-naphtho[2,1-b]pyrano[3,4-e]pyran-3,6-diyl diacetate, CHOLESTEROL, OLEIC ACID, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of PPPA bound human ACAT2
To Be Published
|
|
5EE1
 
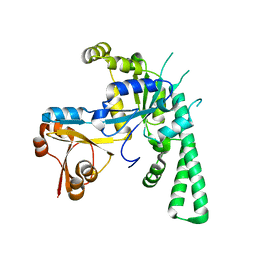 | | Crystal structure of OsYchF1 at pH 7.85 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE3
 
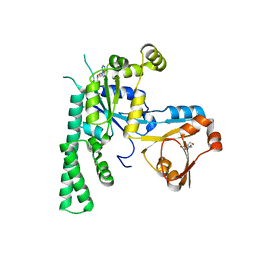 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE0
 
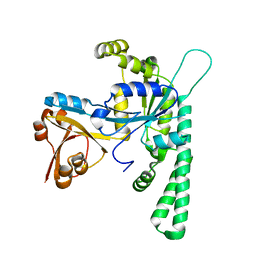 | | Crystal structure of OsYchF1 at pH 6.5 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3NDM
 
 | |
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNW
 
 | | Crystal Structure of the F91STOP mutant of des1-6 Bovine Neurophysin-I, unliganded state | | Descriptor: | Oxytocin-neurophysin 1 | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
4HYD
 
 | | Structure of a presenilin family intramembrane aspartate protease in C2221 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYG
 
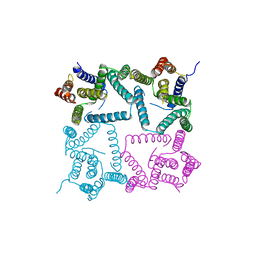 | | Structure of a presenilin family intramembrane aspartate protease in C222 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
3Q8T
 
 | |
3RQF
 
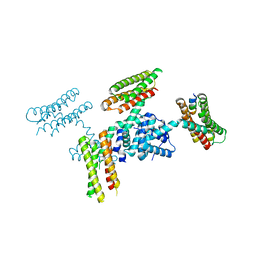 | |
3RQG
 
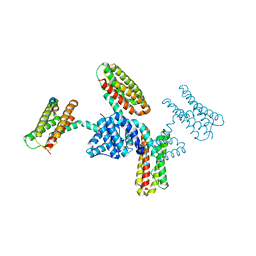 | |
3RQE
 
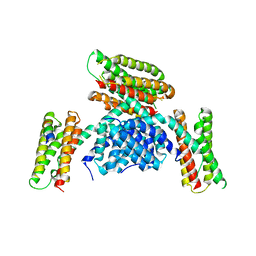 | |
3SIP
 
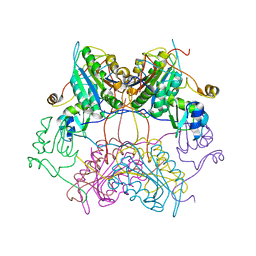 | | Crystal structure of drICE and dIAP1-BIR1 complex | | Descriptor: | Apoptosis 1 inhibitor, Caspase, ZINC ION | | Authors: | Li, X, Wang, J, Shi, Y. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.496 Å) | | Cite: | Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE.
Nat Commun, 2, 2011
|
|
3SIQ
 
 | |
3SIR
 
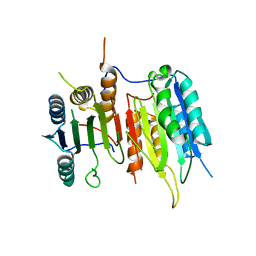 | | Crystal Structure of drICE | | Descriptor: | Caspase | | Authors: | Li, X, Wang, J, Shi, Y. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE.
Nat Commun, 2, 2011
|
|
5YGI
 
 | |
4DXA
 
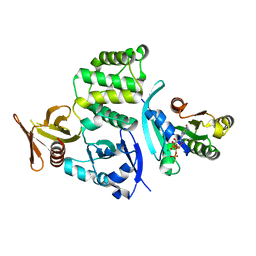 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
5U73
 
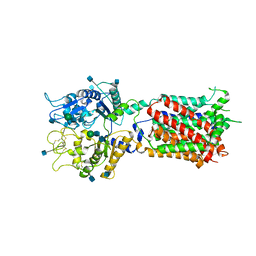 | | Crystal structure of human Niemann-Pick C1 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | Authors: | Li, X, Wang, J, Blobel, G. | | Deposit date: | 2016-12-11 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4EIR
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
5U74
 
 | | Structure of human Niemann-Pick C1 protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4EIS
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, PEROXIDE ION, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
5VB3
 
 | | X-ray structure of nuclear receptor ROR-gammat Ligand Binding Domain + SRC2 peptide | | Descriptor: | Nuclear receptor ROR-gamma, SRC2 chimera, SODIUM ION | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
