3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
3ZIZ
 
 | | Crystal structure of Podospora anserina GH5 beta-(1,4)-mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 ENDO-BETA-1,4-MANNANASE, GLYCEROL | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
6TOP
 
 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
3BE8
 
 | | Crystal structure of the synaptic protein neuroligin 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-01-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and Its beta-Neurexin Complex: Determinants for Folding and Cell Adhesion
Neuron, 56, 2007
|
|
2WQZ
 
 | | Crystal structure of synaptic protein neuroligin-4 in complex with neurexin-beta 1: alternative refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEUREXIN-1-BETA, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and its Beta-Neurexin Complex: Determinants for Folding and Cell Adhesion.
Neuron, 56, 2007
|
|
2V8Q
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V9J
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V92
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
5LZ0
 
 | | Llama nanobody PorM_01 | | Descriptor: | PorM nanobody | | Authors: | Duhoo, Y, Leone, P, Roussel, A. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1TA3
 
 | | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
1TE1
 
 | | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, endo-1,4-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
1WO9
 
 | | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop | | Descriptor: | trypsin inhibitor | | Authors: | Kellenberger, C, Ferrat, G, Leone, P, Darbon, H, Roussel, A. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop
Biochemistry, 42, 2003
|
|
2XZ8
 
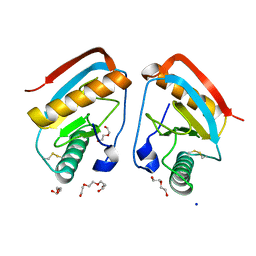 | | CRYSTAL STRUCTURE OF THE LFW ECTODOMAIN OF THE PEPTIDOGLYCAN RECOGNITION PROTEIN LF | | Descriptor: | DI(HYDROXYETHYL)ETHER, PEPTIDOGLYCAN-RECOGNITION PROTEIN LF, SODIUM ION, ... | | Authors: | Basbous, N, Coste, F, Leone, P, Vincentelli, R, Royet, J, Kellenberger, C, Roussel, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Embo Rep., 12, 2011
|
|
2RKQ
 
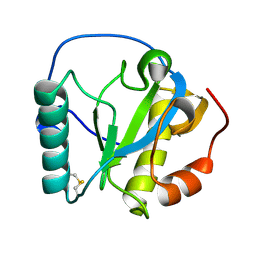 | |
2XXL
 
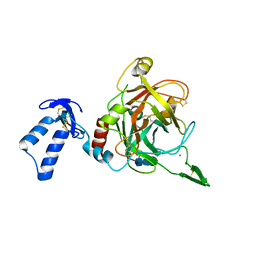 | | Crystal structure of drosophila Grass clip serine protease of Toll pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GRAM-POSITIVE SPECIFIC SERINE PROTEASE, ... | | Authors: | Kellenberger, C, Leone, P, Coquet, L, Jouenne, T, Reichhart, J.M, Roussel, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation.
J.Biol.Chem., 286, 2011
|
|
2XZ4
 
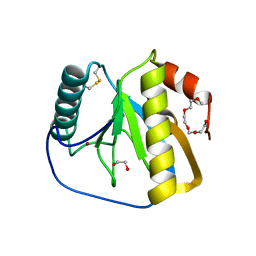 | | Crystal structure of the LFZ ectodomain of the peptidoglycan recognition protein LF | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COPPER (II) ION, ... | | Authors: | Basbous, N, Coste, F, Leone, P, Vincentelli, R, Royet, J, Kellenberger, C, Roussel, A. | | Deposit date: | 2010-11-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Embo Rep., 12, 2011
|
|
7YZW
 
 | |
