8HW9
 
 | | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1
To Be Published
|
|
7Y39
 
 | | Ubiquitin-like domain of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insight into ZFAND1 and p97 Interaction
To Be Published
|
|
7C8S
 
 | | Crystal structure of DUSP22 mutant_N128A | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-06-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
2L8S
 
 | |
2M0Q
 
 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
6L1S
 
 | | Crystal structure of DUSP22 mutant_C88S | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Chang, C.C, Lyu, P.C. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LMY
 
 | | Crystal structure of DUSP22 mutant_C88S/S93A | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2019-12-27 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOT
 
 | | Crystal structure of DUSP22 mutant_N128D | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LVQ
 
 | | Crystal structure of DUSP22_VO4 | | Descriptor: | Dual specificity protein phosphatase 22, VANADATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-02-04 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOU
 
 | | Crystal structure of DUSP22 mutant_C88S/S93N | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5301 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6WCE
 
 | | Structure of the periplasmic binding protein P5PA | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal-5-phosphate binding protein A (P5PA) | | Authors: | Pan, C, Zimmer, A, Shah, M, Huynh, M, Lai, C.C.L, Sit, B, Hooda, Y, Moraes, T.F. | | Deposit date: | 2020-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Actinobacillus utilizes a binding protein-dependent ABC transporter to acquire the active form of vitamin B 6 .
J.Biol.Chem., 297, 2021
|
|
2ACU
 
 | | TYROSINE-48 IS THE PROTON DONOR AND HISTIDINE-110 DIRECTS SUBSTRATE STEREOCHEMICAL SELECTIVITY IN THE REDUCTION REACTION OF HUMAN ALDOSE REDUCTASE: ENZYME KINETICS AND THE CRYSTAL STRUCTURE OF THE Y48H MUTANT ENZYME | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bohren, K.M, Grimshaw, C.E, Lai, C.-J, Gabbay, K.H, Petsko, G.A, Harrison, D.H, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme.
Biochemistry, 33, 1994
|
|
5COQ
 
 | | The effect of valine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-20 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CPF
 
 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CPB
 
 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and inhibition by ligand PT70 (TCU) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CP8
 
 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ... | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
4OHU
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
2M0M
 
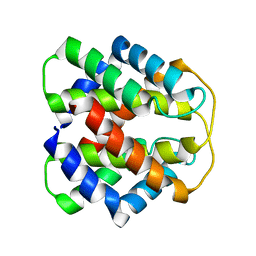 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-22 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7YAB
 
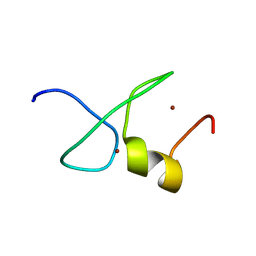 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
4OIM
 
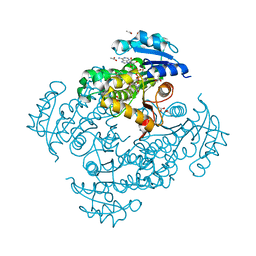 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
7CEM
 
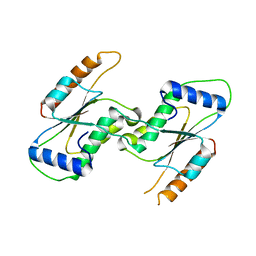 | | Crystal Structure of YbeA CP74 W7F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Wu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4251 Å) | | Cite: | Crystal Structure of YbeA CP74 W7F
To Be Published
|
|
7CFY
 
 | | Crystal Structure of YbeA CP74 W48F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4051 Å) | | Cite: | Crystal Structure of YbeA CP74 W48F
To Be Published
|
|
7CF7
 
 | | Crystal Structure of YbeA CP74 W72F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6178 Å) | | Cite: | Crystal Structure of YbeA CP74 W72F
To Be Published
|
|
7CIU
 
 | |
