2EK7
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M) | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M)
To be Published
|
|
2EKB
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M)
To be Published
|
|
2E8H
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2E8Q
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (K19M) | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Asada, Y, Shimada, H, Taketa, M, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (K19M)
To be Published
|
|
2DXT
 
 | |
2EP5
 
 | |
2EJ9
 
 | |
2EO4
 
 | |
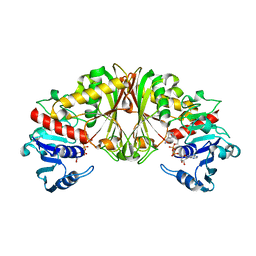
2EP7
 
 | |
2EVB
 
 | |
1J3W
 
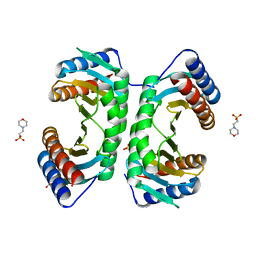 | |
1J2W
 
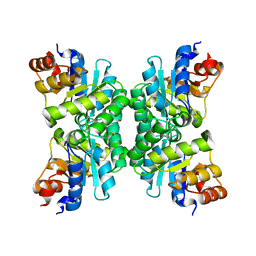 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J3B
 
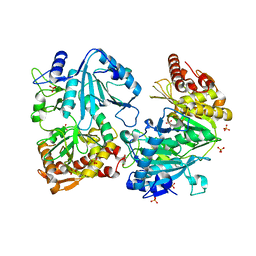 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1HQC
 
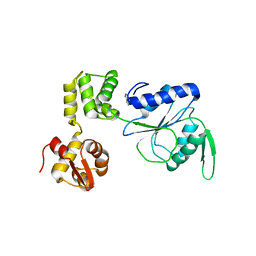 | | STRUCTURE OF RUVB FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | ADENINE, MAGNESIUM ION, RUVB | | Authors: | Yamada, K, Kunishima, N, Mayanagi, K, Iwasaki, H, Morikawa, K. | | Deposit date: | 2000-12-15 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2FYK
 
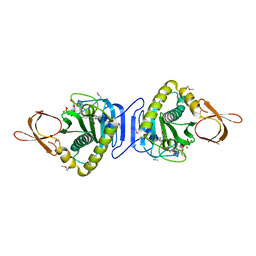 | |
2GJU
 
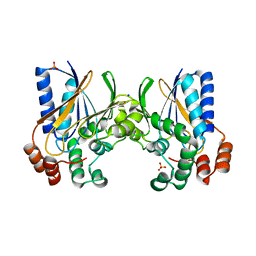 | |
2HD9
 
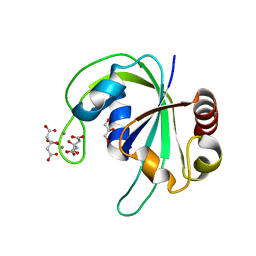 | | Crystal structure of PH1033 from Pyrococcus horikoshii OT3 | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-20 | | Release date: | 2006-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleant-mediated protein crystallization with the application of microporous synthetic zeolites.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2HTM
 
 | |
2HUQ
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | PLATINUM (II) ION, Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sugahara, M, Karthe, P, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2HUN
 
 | |
2HUT
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sugahara, M, Saraboji, K, Malathy sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2HUV
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | PLATINUM (II) ION, Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Sugahara, M, Saraboji, K, Malathy sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2HUX
 
 | |
2HVB
 
 | |
2HR8
 
 | |
