1D53
 
 | | CRYSTAL STRUCTURE AT 1.5 ANGSTROMS RESOLUTION OF D(CGCICICG), AN OCTANUCLEOTIDE CONTAINING INOSINE, AND ITS COMPARISON WITH D(CGCG) AND D(CGCGCG) STRUCTURES | | Descriptor: | DNA (5'-D(*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*IP*CP*IP*CP*G)-3') | | Authors: | Kumar, V.D, Harrison, R.W, Andrews, L.C, Weber, I.T. | | Deposit date: | 1992-11-05 | | Release date: | 1993-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures.
Biochemistry, 31, 1992
|
|
1HCB
 
 | |
1HUH
 
 | |
1HUG
 
 | |
4RMP
 
 | | Crystal structure of allophycocyanin from marine cyanobacterium Phormidium sp. A09DM | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Kumar, V, Gupta, G.D, Sonani, R.R, Madamwar, D. | | Deposit date: | 2014-10-22 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of Allophycocyanin from Marine Cyanobacterium Phormidium sp. A09DM.
Plos One, 10, 2015
|
|
5FVB
 
 | | CRYSTAL STRUCTURE OF PHORMIDIUM C-PHYCOERYTHRIN AT PH 5.0 | | Descriptor: | C-PHYCOERYTHRIN ALPHA SUBUNIT, C-PHYCOERYTHRIN BETA SUBUNIT, GLYCEROL, ... | | Authors: | Kumar, V, Sonani, R.R, Sharma, M, Gupta, G.D, Madamwar, D. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
5IXX
 
 | |
5IXR
 
 | |
5IXW
 
 | |
5IXV
 
 | |
5IXZ
 
 | |
3UH1
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
4DG7
 
 | |
3TUY
 
 | | Phosphorylated Light Chain Domain of Scallop smooth Muscle Myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Kumar, V.S.S, O'Neall-hennessey, E, Reshetnikova, L, Brown, J.H, Robinson, H, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2011-09-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Biophys.J., 101, 2011
|
|
5AQD
 
 | | Crystal structure of Phormidium Phycoerythrin at pH 8.5 | | Descriptor: | GLYCEROL, PHYCOERYTHRIN ALPHA SUBUNIT, PHYCOERYTHRIN BETA SUBUNIT, ... | | Authors: | Kumar, V, Sharma, M, Sonani, R.R, Gupta, G.D, Madamwar, D. | | Deposit date: | 2015-09-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
6K5P
 
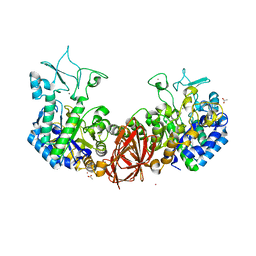 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
4E53
 
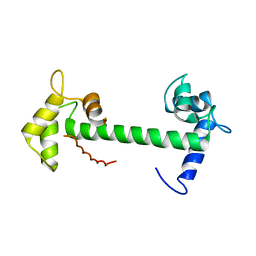 | | Calmodulin and Nm peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neuromodulin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
4E50
 
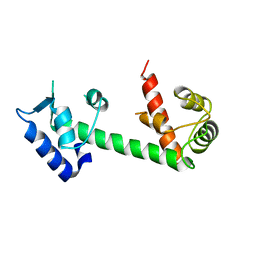 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
3TS5
 
 | | Crystal Structure of a Light Chain Domain of Scallop Smooth Muscle Myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Kumar, V.S.S, O'Neall-Hennessey, E, Reshetnikova, L, Brown, J.H, Robinson, H, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2011-09-12 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Biophys.J., 101, 2011
|
|
4HEX
 
 | |
3T0I
 
 | |
3T7S
 
 | |
3SXJ
 
 | |
4KQN
 
 | |
4KIR
 
 | |
