1JMW
 
 | |
9ICD
 
 | | CATALYTIC MECHANISM OF NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE: IMPLICATIONS FROM THE STRUCTURES OF MAGNESIUM-ISOCITRATE AND NADP+ COMPLEXES | | Descriptor: | ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hurley, J.H, Dean, A.M, Koshland Jr, D.E, Stroud, R.M. | | Deposit date: | 1991-07-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes.
Biochemistry, 30, 1991
|
|
2AGG
 
 | | succinyl-AAPK-trypsin acyl-enzyme at 1.28 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AGI
 
 | | The leupeptin-trypsin covalent complex at 1.14 A resolution | | Descriptor: | CALCIUM ION, SULFATE ION, beta-trypsin, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AGE
 
 | | Succinyl-AAPR-trypsin acyl-enzyme at 1.15 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin, succinyl-Ala-Ala-Pro-Arg | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AH4
 
 | | guanidinobenzoyl-trypsin acyl-enzyme at 1.13 A resolution | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, SULFATE ION, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1PB1
 
 | |
1PB3
 
 | |
1LW6
 
 | |
1Y3C
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 R62A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y3B
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 E60S mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y48
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 R65A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y33
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 T58P mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-23 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y4A
 
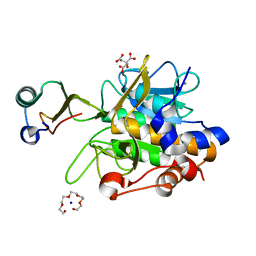 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59R/E60S mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y34
 
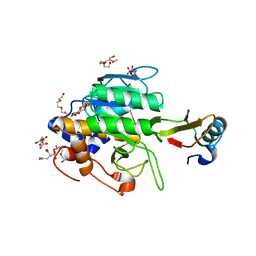 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 E60A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-23 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y4D
 
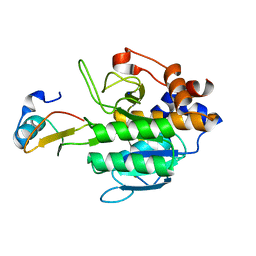 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59R/E60S mutant | | Descriptor: | CALCIUM ION, SODIUM ION, chymotrypsin inhibitor 2, ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y1K
 
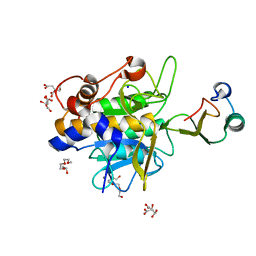 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 T58A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y3F
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 F69A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1Y3D
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 R67A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
1OYE
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY9
 
 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, ETHIDIUM | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OYD
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, DEQUALINIUM | | Authors: | Yu, E.W, MeDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY6
 
 | | Structural Basis of the Multiple Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY8
 
 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, RHODAMINE 6G | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1P8F
 
 | |
