3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
6KSL
 
 | | Staphylococcus aureus lipase - S116A inactive mutant | | Descriptor: | CALCIUM ION, LAURIC ACID, Lipase 2, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
1G8Q
 
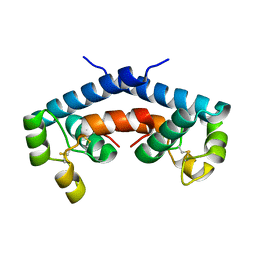 | | CRYSTAL STRUCTURE OF HUMAN CD81 EXTRACELLULAR DOMAIN, A RECEPTOR FOR HEPATITIS C VIRUS | | Descriptor: | CD81 ANTIGEN, EXTRACELLULAR DOMAIN | | Authors: | Kitadokoro, K, Bolognesi, M, Bordo, D, Grandi, G, Galli, G, Petracca, R, Falugi, F. | | Deposit date: | 2000-11-20 | | Release date: | 2001-02-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
EMBO J., 20, 2001
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
2SFA
 
 | | SERINE PROTEINASE FROM STREPTOMYCES FRADIAE ATCC 14544 | | Descriptor: | SERINE PROTEINASE | | Authors: | Kitadokoro, K, Tsuzuki, H. | | Deposit date: | 1994-04-25 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, characterization, primary structure, crystallization and preliminary crystallographic study of a serine proteinase from Streptomyces fradiae ATCC 14544.
Eur.J.Biochem., 220, 1994
|
|
3VIS
 
 | | Crystal structure of cutinase Est119 from Thermobifida alba AHK119 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Esterase | | Authors: | Kitadokoro, K, Thumarat, U, Nakamura, R, Nishimura, K, Karatani, H, Suzuki, H, Kawai, F. | | Deposit date: | 2011-10-11 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
POLYM.DEGRAD.STAB., 97, 2012
|
|
6KSM
 
 | | Staphylococcus aureus lipase -Orlistat complex | | Descriptor: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, CALCIUM ION, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
6KSI
 
 | | Staphylococcus aureus lipase - native | | Descriptor: | CALCIUM ION, HEXANOIC ACID, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
1IV5
 
 | | New Crystal Form of Human CD81 Large Extracellular Loop. | | Descriptor: | CD81 antigen | | Authors: | Kitadokoro, K, Bolognesi, M, Grandi, G, Marco, P, Galli, G, Petracca, R, Fabiana, F. | | Deposit date: | 2002-03-14 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Subunit Association and Conformational Flexibility in the Head-subdomain of Human CD81 Large Extracellular Loop.
Biol.Chem., 383, 2002
|
|
2EC5
 
 | |
2EBF
 
 | |
2EBH
 
 | |
3WYN
 
 | |
1QNF
 
 | | STRUCTURE OF PHOTOLYASE | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, FLAVIN-ADENINE DINUCLEOTIDE, PHOTOLYASE | | Authors: | Miki, K, Kitadokoro, K. | | Deposit date: | 1997-07-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA photolyase from Anacystis nidulans
Nat.Struct.Biol., 4, 1997
|
|
3WIN
 
 | | Clostridium botulinum Hemagglutinin | | Descriptor: | 17 kD hemagglutinin component, HA1, HA3 | | Authors: | Amatsu, S, Sugawara, Y, Matsumura, T, Fujinaga, Y, Kitadokoro, K. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Clostridium botulinum Whole Hemagglutinin Reveals a Huge Triskelion-shaped Molecular Complex
J.Biol.Chem., 288, 2013
|
|
