2CVO
 
 | | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) | | Descriptor: | putative Semialdehyde dehydrogenase | | Authors: | Nonaka, T, Kita, A, Miura-Ohnuma, J, Katoh, E, Inagaki, N, Yamazaki, T, Miki, K. | | Deposit date: | 2005-06-10 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa)
Proteins, 61, 2005
|
|
2YQC
 
 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2ZFO
 
 | | Structure of the partially unliganded met state of 400 kDa hemoglobin: Insights into ligand-induced structural changes of giant hemoglobins | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B1, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the partially unliganded met state of 400 kDa hemoglobin: insights into ligand-induced structural changes of giant hemoglobins
Proteins, 73, 2008
|
|
2ZHG
 
 | | Crystal structure of SoxR in complex with DNA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZHH
 
 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZS0
 
 | | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular giant hemoglobin major globin subunit A1, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin
Biochemistry, 47, 2008
|
|
2ZS1
 
 | | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin | | Descriptor: | CHLORIDE ION, Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin
Biochemistry, 47, 2008
|
|
3WCU
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Deoxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCV
 
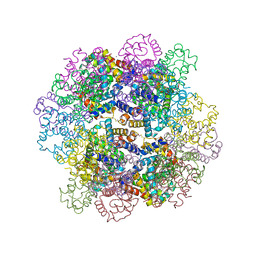 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: CA bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCW
 
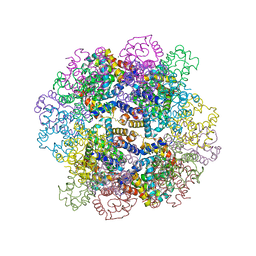 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: MG bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCT
 
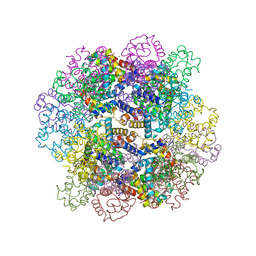 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Oxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WDL
 
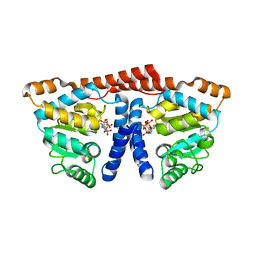 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with ATP | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WDK
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with reaction intermediate | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, 5'-O-[(S)-hydroxy{[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]oxy}phosphoryl]adenosine, CITRATE ANION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WDM
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase from Thermococcus kodakarensis | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
