8GN5
 
 | | Designed pH-responsive P22 VLP | | Descriptor: | Major capsid protein | | Authors: | Kim, K.J, Kim, G, Bae, J.H, Song, J.J, Kim, H.S. | | Deposit date: | 2022-08-23 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A pH-Responsive Virus-Like Particle as a Protein Cage for a Targeted Delivery.
Adv Healthc Mater, 13, 2024
|
|
2CB0
 
 | |
8JB1
 
 | |
4WYS
 
 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4WYR
 
 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL4
 
 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL2
 
 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL3
 
 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
8JZH
 
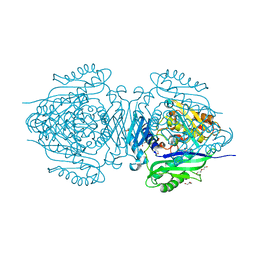 | | C. glutamicum S-adenosylmethionine synthase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, S-adenosylmethionine synthase, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZI
 
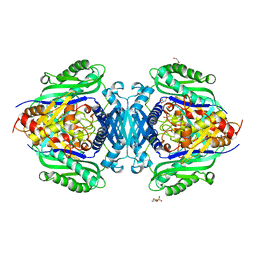 | | Mutant S-adenosylmethionine synthase from C. glutamicum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZG
 
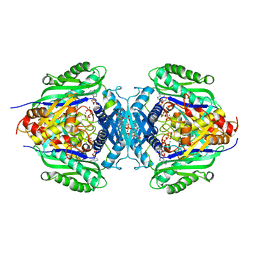 | | C. glutamicum S-adenosylmethionine synthase co-crystallized with Adenosine, triphosphate, and SAM | | Descriptor: | ADENOSINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8HRQ
 
 | |
8HRT
 
 | |
8HRP
 
 | |
8HRR
 
 | |
8HRO
 
 | |
8HRS
 
 | |
2WL9
 
 | | Crystal structure of catechol 2,3-dioxygenase | | Descriptor: | 3-METHYLCATECHOL, CATECHOL 2,3-DIOXYGENASE, FE (III) ION, ... | | Authors: | Cho, H.J, Kim, K.J, Kang, B.S. | | Deposit date: | 2009-06-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-Binding Mechanism of a Type I Extradiol Dioxygenase.
J.Biol.Chem., 285, 2010
|
|
2WL3
 
 | | crystal structure of catechol 2,3-dioxygenase | | Descriptor: | CALCIUM ION, CATECHOL 2,3-DIOXYGENASE, FE (III) ION, ... | | Authors: | Cho, H.J, Kim, K.J, Kang, B.S. | | Deposit date: | 2009-06-21 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-Binding Mechanism of a Type I Extradiol Dioxygenase.
J.Biol.Chem., 285, 2010
|
|
2BR6
 
 | | Crystal Structure of Quorum-Quenching N-Acyl Homoserine Lactone Lactonase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, HOMOSERINE LACTONE, ... | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Kang, B.S, Kim, K.J, Derewenda, Z.S, Lee, J.K, Oh, T.K, Lee, C.H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BTN
 
 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6IJ6
 
 | |
6IJ3
 
 | |
6IJ5
 
 | |
6IUN
 
 | |
