7Y2R
 
 | |
7Y2M
 
 | |
7Y2U
 
 | |
7Y2I
 
 | |
7Y2O
 
 | |
7Y2G
 
 | |
7Y2K
 
 | |
7Y2N
 
 | |
7Y2W
 
 | |
7Y2J
 
 | |
7Y2Q
 
 | |
2LN4
 
 | |
4NT8
 
 | | Formyl-methionine-alanine complex structure of peptide deformylase from Xanthomoonas oryzae pv. oryzae | | Descriptor: | ACETATE ION, ALANINE, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-12-02 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure of Xoo1075, a peptide deformylase, from Xanthomonas oryzae pv. oryzae
To be Published
|
|
3DKU
 
 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3HE8
 
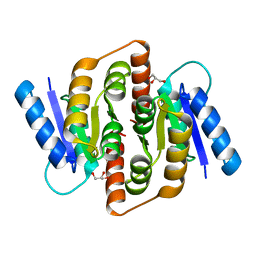 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3HEE
 
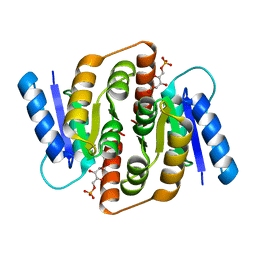 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B and ribose-5-phosphate | | Descriptor: | RIBOSE-5-PHOSPHATE, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3PH3
 
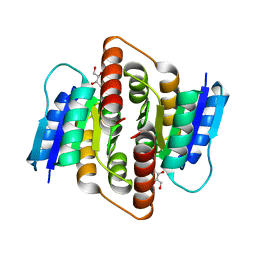 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-ribose | | Descriptor: | D-ribose, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.K, Yeom, S.J, Ahn, Y.J, Oh, D.K, Kang, L.W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
4IXZ
 
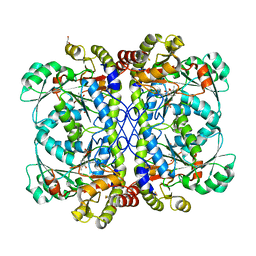 | | Native structure of cystathionine gamma lyase (XometC) from xanthomonas oryzae pv. oryzae at pH 9.0 | | Descriptor: | BETA-MERCAPTOETHANOL, BICARBONATE ION, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IYO
 
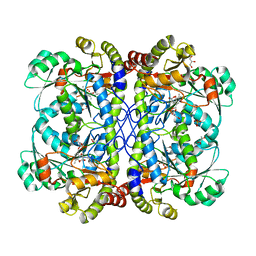 | | Crystal structure of cystathionine gamma lyase from Xanthomonas oryzae pv. oryzae (XometC) in complex with E-site serine, A-site serine, A-site external aldimine structure with aminoacrylate and A-site iminopropionate intermediates | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMINO-ACRYLATE, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IXS
 
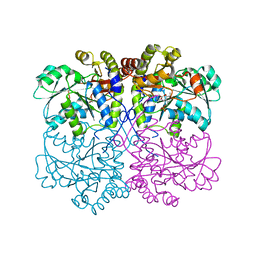 | | Native structure of xometc at ph 5.2 | | Descriptor: | CARBONATE ION, Cystathionine gamma-lyase-like protein, GLYCEROL | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IY7
 
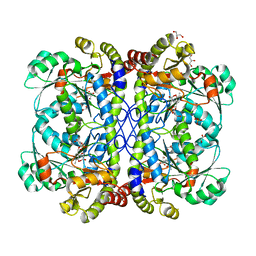 | | crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L1K
 
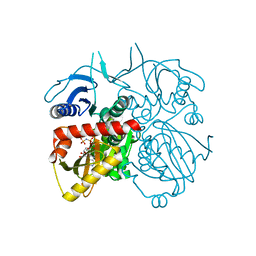 | |
3RFC
 
 | |
3R5F
 
 | |
4ME6
 
 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
