6HH2
 
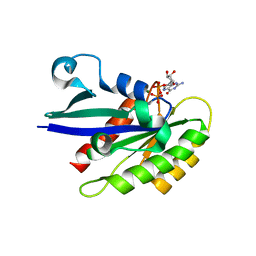 | | Rab29 small GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7L1 | | Authors: | Khan, A.R, McGrath, E, Waschbusch, D. | | Deposit date: | 2018-08-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | LRRK2 binds to the Rab32 subfamily in a GTP-dependent mannerviaits armadillo domain.
Small GTPases, 2019
|
|
2GZD
 
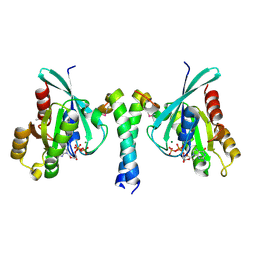 | | Crystal Structure of Rab11 in Complex with Rab11-FIP2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab11 family-interacting protein 2, ... | | Authors: | Khan, A.R. | | Deposit date: | 2006-05-11 | | Release date: | 2006-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of rab11 in complex with rab11 family interacting protein 2.
Structure, 14, 2006
|
|
2GZH
 
 | |
7KPR
 
 | |
7L4I
 
 | |
7L4J
 
 | | Crystal structure of WT PPM1H phosphatase | | Descriptor: | MAGNESIUM ION, Protein phosphatase 1H | | Authors: | Khan, A.R, Waschbusch, D. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural basis for the specificity of PPM1H phosphatase for Rab GTPases.
Embo Rep., 22, 2021
|
|
6ETC
 
 | |
6FF8
 
 | |
6SQ2
 
 | |
1DUZ
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201) IN COMPLEX WITH A NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1DUY
 
 | | CRYSTAL STRUCTURE OF HLA-A*0201/OCTAMERIC TAX PEPTIDE COMPLEX | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A2*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1AVF
 
 | | ACTIVATION INTERMEDIATE 2 OF HUMAN GASTRICSIN FROM HUMAN STOMACH | | Descriptor: | GASTRICSIN, SODIUM ION | | Authors: | Khan, A.R, Cherney, M.M, Tarasova, N.I, James, M.N.G. | | Deposit date: | 1997-09-16 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural characterization of activation 'intermediate 2' on the pathway to human gastricsin.
Nat.Struct.Biol., 4, 1997
|
|
7N0Z
 
 | |
6WHE
 
 | |
8OQH
 
 | |
3TYI
 
 | | Crystal Structure of Cytochrome c - p-Sulfonatocalix[4]arene Complexes | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mc Govern, R.E, Fernandes, H, Khan, A.R, Crowley, P.B. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Protein camouflage in cytochrome c-calixarene complexes.
Nat Chem, 4, 2012
|
|
3TSO
 
 | | Structure of the cancer associated Rab25 protein in complex with FIP2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Oda, S, Lall, P.Y, McCaffrey, M.W, Khan, A.R. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cancer associated Rab25 protein in complex with FIP2
TO BE PUBLISHED
|
|
7P53
 
 | |
3CWZ
 
 | | Structure of RAB6(GTP)-R6IP1 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab6-interacting protein 1, ... | | Authors: | Recacha, R, Houdusse, A, Goud, B, Khan, A.R. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain.
Structure, 17, 2009
|
|
7U0V
 
 | |
7U0W
 
 | |
2W8C
 
 | |
2W88
 
 | |
8GII
 
 | |
