6IKT
 
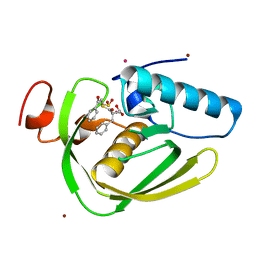 | |
6IL0
 
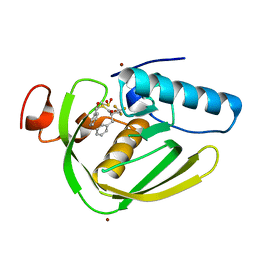 | |
6IL9
 
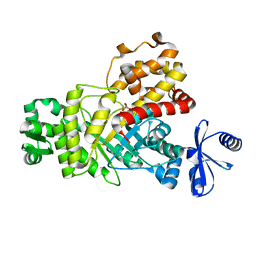 | | One Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, GLYCEROL, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72005355 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To Be Published
|
|
6IKY
 
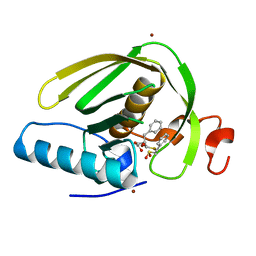 | |
5C1O
 
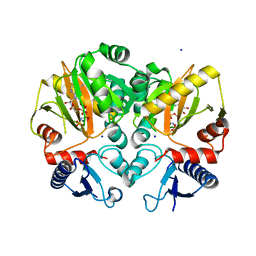 | | Crystal structure of AMP-PNP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5C1P
 
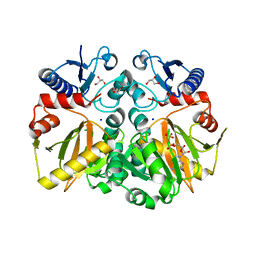 | | Crystal structure of ADP and D-alanyl-D-alanine complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K, Ngo, H.P.T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6JFR
 
 | |
6JET
 
 | |
6JFO
 
 | |
6JF3
 
 | |
6JES
 
 | |
6JEW
 
 | |
6JF9
 
 | |
6JFC
 
 | |
6JF4
 
 | |
6JFF
 
 | |
6JFQ
 
 | |
6JFA
 
 | |
6JER
 
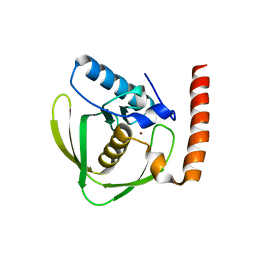 | | Apo crystal structure of class I type a peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, crystallization, and preliminary X-ray crystallographic analysis of peptide deformylase from Acinetobacter baumanii
Biodesign, 5, 2017
|
|
6JEU
 
 | |
6JF7
 
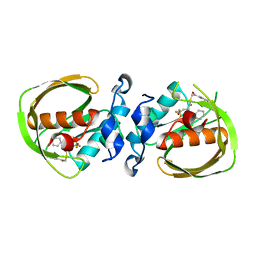 | | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, S-(2-oxo-2-phenylethyl) (2R)-2-benzyl-4,4,4-trifluorobutanethioate, ZINC ION | | Authors: | Jung, K.H, Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-08 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii
To be published
|
|
6JEV
 
 | |
6JFG
 
 | |
6JF5
 
 | |
6JFE
 
 | |
