1J1F
 
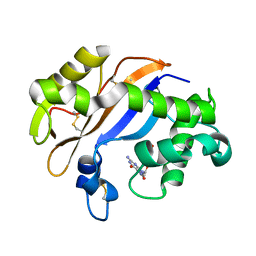 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
3VKK
 
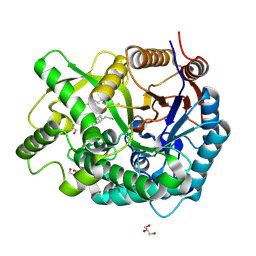 | | Crystal Structure Of The Covalent Intermediate Of Human Cytosolic Beta-Glucosidase-mannose complex | | Descriptor: | CHLORIDE ION, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Okino, N, Ito, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition mechanism of human cytosolic beta-glucosidase by monnoside
To be Published
|
|
5WRI
 
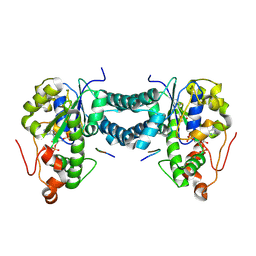 | | Crystal structure of human tyrosylprotein sulfotransferase-1 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ASP-PHE-GLU-ASP-TYR-GLU-PHE-ASP, GLYCEROL, ... | | Authors: | Tanaka, S, Nishiyori, T, Kojo, H, Otsubo, R, Kakuta, Y. | | Deposit date: | 2016-12-02 | | Release date: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.
Sci Rep, 7, 2017
|
|
1UAR
 
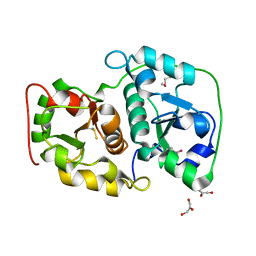 | |
1CJM
 
 | | HUMAN SULT1A3 WITH SULFATE BOUND | | Descriptor: | PROTEIN (ARYL SULFOTRANSFERASE), SULFATE ION | | Authors: | Bidwell, L.M, Mcmanus, M.E, Gaedigk, A, Kakuta, Y, Negishi, M, Pedersen, L, Martin, J.L. | | Deposit date: | 1999-04-18 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human catecholamine sulfotransferase.
J.Mol.Biol., 293, 1999
|
|
2CZW
 
 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2CZV
 
 | | Crystal structure of archeal RNase P protein ph1481p in complex with ph1877p | | Descriptor: | ACETIC ACID, Ribonuclease P protein component 2, Ribonuclease P protein component 3, ... | | Authors: | Kawano, S, Kakuta, Y, Nakashima, T, Tanaka, I, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-06-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme
J.Mol.Biol., 357, 2006
|
|
6KZR
 
 | | Crystal structure of mouse DCAR2 CRD domain | | Descriptor: | C-type lectin domain family 4, member b1, CALCIUM ION, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Teramoto, T, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
6LFJ
 
 | | Crystal structure of mouse DCAR2 CRD domain complex with IPM2 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
6LKR
 
 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
5X2B
 
 | |
5Z8B
 
 | |
1IYB
 
 | |
3VU2
 
 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltopentaose from Oryza sativa L | | Descriptor: | 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, GLYCEROL, ... | | Authors: | Chaen, K, Kakuta, Y, Kimura, M. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the rice branching enzyme I (BEI) in complex with maltopentaose.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
3WZ0
 
 | | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | Ribonuclease P protein component 2, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WYZ
 
 | | On archaeal homologs of the human RNase P protein Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | GLYCEROL, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
7EOV
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT2A8 in complex with PAP and cholic acid | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CHOLIC ACID, cytosolic sulfotransferase SULT2A8 | | Authors: | Teramoto, T, Nishio, T, Kakuta, Y. | | Deposit date: | 2021-04-22 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mouse SULT2A8 reveals the mechanism of 7 alpha-hydroxyl, bile acid sulfation.
Biochem.Biophys.Res.Commun., 562, 2021
|
|
2ZOX
 
 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
1UAX
 
 | |
1V9H
 
 | | Crystal structure of the RNase MC1 mutant Y101A in complex with 5'-UMP | | Descriptor: | Ribonuclease MC, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kimura, K, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-01-26 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino acids conserved at the C-terminal half of the ribonuclease t2 family contribute to protein stability of the enzymes
Biosci.Biotechnol.Biochem., 68, 2004
|
|
1V77
 
 | | Crystal structure of the PH1877 protein | | Descriptor: | hypothetical protein PH1877 | | Authors: | Takagi, H, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1V76
 
 | |
1VD1
 
 | |
1VD3
 
 | | Ribonuclease NT in complex with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, RNase NGR3 | | Authors: | Kawano, S, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-03-18 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Nicotiana glutinosa Ribonuclease NT in Complex with Nucleotide Monophosphates
to be published
|
|
1VCZ
 
 | |
