1ZC3
 
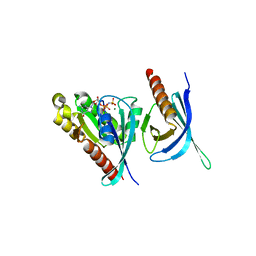 | | Crystal structure of the Ral-binding domain of Exo84 in complex with the active RalA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-A, ... | | Authors: | Jin, R, Junutula, J.R, Matern, H.T, Ervin, K.E, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2005-04-10 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase.
Embo J., 24, 2005
|
|
1ZC4
 
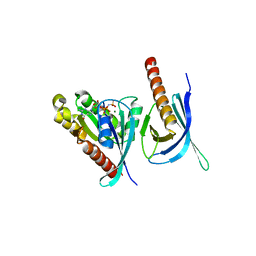 | | Crystal structure of the Ral-binding domain of Exo84 in complex with the active RalA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-A, ... | | Authors: | Jin, R, Junutula, J.R, Matern, H.T, Ervin, K.E, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2005-04-10 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase.
Embo J., 24, 2005
|
|
3SAJ
 
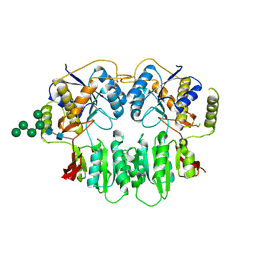 | | Crystal Structure of glutamate receptor GluA1 Amino Terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 1, ... | | Authors: | Jin, R, Zong, Y, Yao, G, Gu, S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the glutamate receptor GluA1 N-terminal domain.
Biochem.J., 438, 2011
|
|
6BVD
 
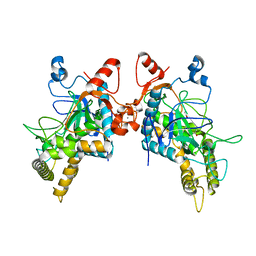 | | Structure of Botulinum Neurotoxin Serotype HA Light Chain | | Descriptor: | ACETATE ION, CALCIUM ION, Light Chain, ... | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA.
Pathog Dis, 76, 2018
|
|
7UIE
 
 | | Crystal structure of HcE-JLE-G6 | | Descriptor: | Botulinum neurotoxin E heavy chain, JLE-G6 | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
2NM1
 
 | | Structure of BoNT/B in complex with its protein receptor | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-2 | | Authors: | Jin, R, Rummel, A, Binz, T, Brunger, A.T. | | Deposit date: | 2006-10-20 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity.
Nature, 444, 2006
|
|
6UFT
 
 | |
6UL6
 
 | |
6UI1
 
 | | Crystal structure of BoNT/A-LCHn domain in complex with VHH ciA-D12, ciA-B5, and ciA-H7 | | Descriptor: | BoNT/A, ciA-B5, ciA-D12, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2019-09-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.20000863 Å) | | Cite: | Structural Insights into Rational Design of Single-Domain Antibody-Based Antitoxins against Botulinum Neurotoxins
Cell Rep, 30, 2020
|
|
6UC6
 
 | |
6UL4
 
 | |
6UHT
 
 | |
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
7ML7
 
 | |
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
7NA9
 
 | | Crystal structure of BoNT/B-LC-JSG-C1 | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type B, JSG-C1, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
6OQ5
 
 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQ6
 
 | |
6OQ8
 
 | |
6OQ7
 
 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6DKK
 
 | | Structure of BoNT | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
4LO3
 
 | | HA17-HA33-LacNac | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
