5JHS
 
 | |
5FG9
 
 | | Yeast 20S proteasome beta2-T(-2)V mutant | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-12-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5FHS
 
 | |
5FGF
 
 | |
5FG7
 
 | | Yeast 20S proteasome beta2-T1A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-12-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5FGG
 
 | |
5FGD
 
 | |
5FGI
 
 | |
7AW9
 
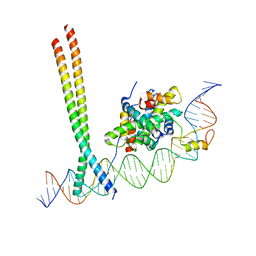 | | CCAAT-binding complex and HapX bound to Aspergillus fumigatus cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
7AW7
 
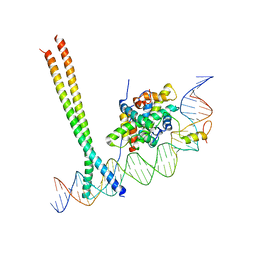 | | CCAAT-binding complex and HapX bound to Aspergillus nidulans cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
5MSK
 
 | | Mouse PA28beta | | Descriptor: | Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
5LWZ
 
 | | Cys-Gly dipeptidase GliJ (space group C2) | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX4
 
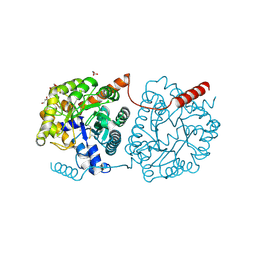 | | Cys-Gly dipeptidase GliJ mutant D38H | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX0
 
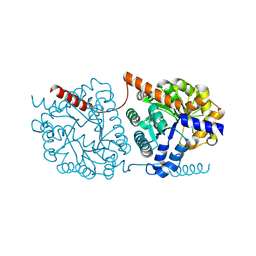 | | Cys-Gly dipeptidase GliJ (space group P3221) | | Descriptor: | Dipeptidase, FE (III) ION | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5MSJ
 
 | | Mouse PA28alpha | | Descriptor: | Proteasome activator complex subunit 1 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
5MX5
 
 | | Mouse PA28alpha-beta | | Descriptor: | PHOSPHATE ION, Proteasome activator complex subunit 1, Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-20 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
5FGH
 
 | |
5JHR
 
 | | Yeast 20S proteasome in complex with the peptidic epoxyketone inhibitor 27 | | Descriptor: | (2S)-2-azido-N-[(2S)-3-(biphenyl-4-yl)-1-{[(2S)-1-{[(2S,3S,4R)-3,5-dihydroxy-4-methylpentan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl]amino}-1-oxopropan-2-yl]-3-phenylpropanamide (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-04-21 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of beta 5c Selective Inhibitors of Human Constitutive Proteasomes.
J.Med.Chem., 59, 2016
|
|
8BII
 
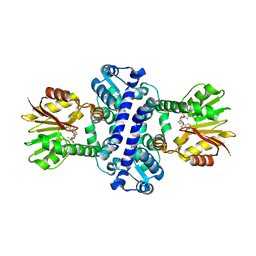 | | O-Methyltransferase Plu4895 (mutant H229N) in complex with SAH | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase Plu4895 H229N mutant | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIE
 
 | | O-Methyltransferase Plu4894 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase Plu4894 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BGX
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-270a | | Descriptor: | 1-methoxy-3,8-bis(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BGT
 
 | | O-Methyltransferase Plu4890 in complex with SAM | | Descriptor: | GLYCEROL, Methyltransferase Plu4890, S-ADENOSYLMETHIONINE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIJ
 
 | | O-Methyltransferase Plu4894 (mutant I88M, W91L, C97Y, S142L, G146V, Y258M, L270F, S309Y) in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Methyltransferase Plu4894 mutant I88M, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIC
 
 | | O-Methyltransferase Plu4891 in complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIR
 
 | | O-Methyltransferase Plu4895 in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, IODIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
