4MQ9
 
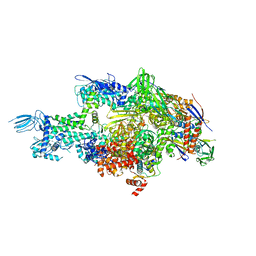 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
3U96
 
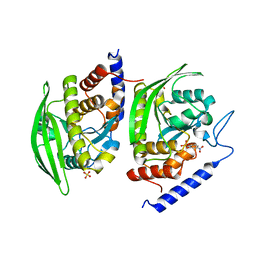 | | Crystal Structure of YopHQ357F(Catalytic Domain, Residues 163-468) in complex with pNCS | | Descriptor: | N,4-DIHYDROXY-N-OXO-3-(SULFOOXY)BENZENAMINIUM, SULFATE ION, Tyrosine-protein phosphatase yopH | | Authors: | Ho, M.C, Ke, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of catalytic loop structure, dynamics, and function relationship of Yersinia protein tyrosine phosphatase by temperature-jump relaxation spectroscopy and X-ray structural determination.
J.Phys.Chem.B, 116, 2012
|
|
4GBD
 
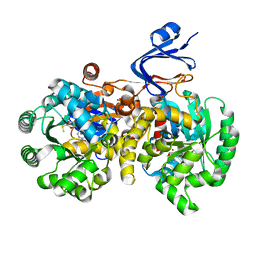 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
4G56
 
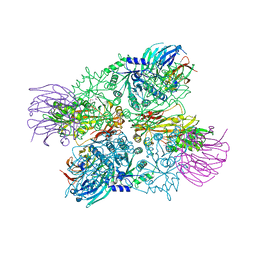 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
3LOO
 
 | | Crystal structure of Anopheles gambiae adenosine kinase in complex with P1,P4-di(adenosine-5) tetraphosphate | | Descriptor: | Anopheles gambiae adenosine kinase, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ho, M.-C, Cassera, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A High-Affinity Adenosine Kinase from Anopheles gambiae.
Biochemistry, 50, 2011
|
|
3HIQ
 
 | |
3HIV
 
 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIT
 
 | | Crystal structure of Saporin-L1 in complex with the dinucleotide inhibitor, a transition state analogue | | Descriptor: | 5'-O-[(S)-{[(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[(S)-hydroxy(3-hydroxypropoxy)phosphoryl]oxy}methyl)pyrrolidin-3-yl]oxy}(hydroxy)phosphoryl]-3'-O-[(R)-hydroxy(4-hydroxybutoxy)phosphoryl]-2'-O-methylguanosine, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIO
 
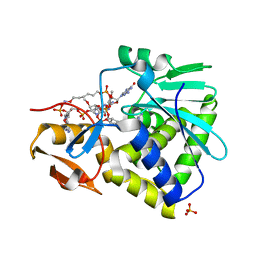 | | Crystal structure of Ricin A-chain in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Ricin, SULFATE ION | | Authors: | Ho, M, Sturm, M.B, Goldman, J.D, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIW
 
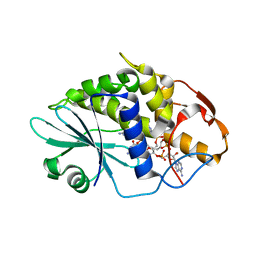 | | Crystal structure of Saporin-L1 in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIS
 
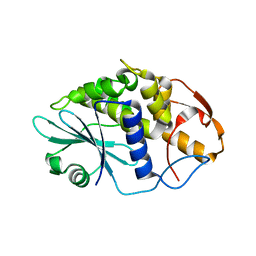 | |
3MB8
 
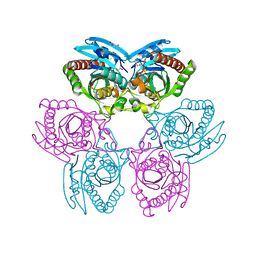 | | Crystal structure of purine nucleoside phosphorylase from toxoplasma gondii in complex with immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ho, M, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Eukaryot Cell, 13, 2014
|
|
3OZG
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with S-SerMe-ImmH phosphonate | | Descriptor: | Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
3OZF
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
7CN9
 
 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
3OZB
 
 | | Crystal Structure of 5'-methylthioinosine phosphorylase from Psedomonas aeruginosa in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylthioinosine phosphorylase from Pseudomonas aeruginosa. Structure and annotation of a novel enzyme in quorum sensing.
Biochemistry, 50, 2011
|
|
4EB8
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
5YTA
 
 | |
3DHF
 
 | | Crystal structure of phosphorylated mimic form of human NAMPT complexed with nicotinamide mononucleotide and pyrophosphate | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-17 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DKL
 
 | | Crystal structure of phosphorylated mimic form of human NAMPT complexed with benzamide and phosphoribosyl pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BENZAMIDE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-25 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K8O
 
 | | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-ImmH | | Descriptor: | 7-({[(1R,2S)-2,3-DIHYDROXY-1-(HYDROXYMETHYL)PROPYL]AMINO}METHYL)-3,5-DIHYDRO-4H-PYRROLO[3,2-D]PYRIMIDIN-4-ONE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Ho, M, Rinaldo-matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-Immucillin H
Proc.Natl.Acad.Sci.USA, 2010
|
|
3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3PHB
 
 | | Crystal Structure of human purine nucleoside phosphorylase in complex with DADMe-ImmG | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Cassera, M.B, Murkin, A.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-11-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of human purine nucleoside phosphorylase in complex with DADMe-ImmG
to be published
|
|
3DGR
 
 | | Crystal structure of human NAMPT complexed with ADP analogue | | Descriptor: | Nicotinamide phosphoribosyltransferase, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DHD
 
 | | Crystal structure of human NAMPT complexed with nicotinamide mononucleotide and pyrophosphate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-17 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
