6S5Q
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-isobutyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-(2-methylpropyl)-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5M
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-n-propyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-propyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
2UZ1
 
 | | 1.65 Angstrom structure of Benzaldehyde Lyase complexed with 2-methyl- 2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZALDEHYDE LYASE, THIAMINE DIPHOSPHATE | | Authors: | Maraite, A, Schmidt, T, Ansorge-Schumacher, M.B, Brzozowski, A.M, Grogan, G. | | Deposit date: | 2007-04-23 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Thdp-Dependent Enzyme Benzaldehyde Lyase Refined to 1.65 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2VVM
 
 | | The structure of MAO-N-D5, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N, ... | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
2VSS
 
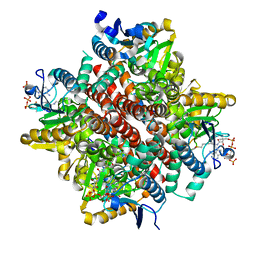 | | Wild-type Hydroxycinnamoyl-CoA hydratase lyase in complex with acetyl- CoA and vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
2VSU
 
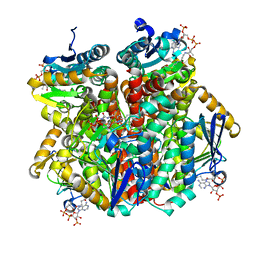 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
5A4V
 
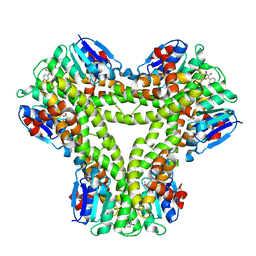 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A4U
 
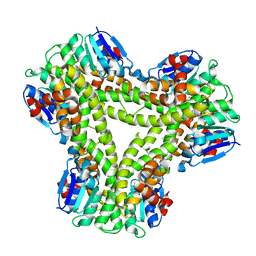 | | AtGSTF2 from Arabidopsis thaliana in complex with indole-3-aldehyde | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A4W
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
6SKX
 
 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
4C5O
 
 | | Flavin monooxygenase from Stenotrophomonas maltophilia. Q193R H194T mutant | | Descriptor: | FLAVIN MONOOXYGENASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2013-09-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutations of an Nad(P)H-Dependent Flavoprotein Monooxygenase that Influence Cofactor Promiscuity and Enantioselectivity.
FEBS Open Bio, 3, 2013
|
|
3ZGJ
 
 | |
4A6G
 
 | | N-acyl amino acid racemase from Amycalotopsis sp. Ts-1-60: G291D- F323Y mutant in complex with N-acetyl methionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-ACYLAMINO ACID RACEMASE | | Authors: | Baxter, S, Royer, S, Grogan, G, Holt-Tiffin, K.E, Taylor, I.N, Fotheringham, I.G, Campopiano, D.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An Improved Racemase/Acylase Biotransformation for the Preparation of Enantiomerically Pure Amino Acids.
J.Am.Chem.Soc., 134, 2012
|
|
4ATQ
 
 | | GABA-transaminase A1R958 in complex with external aldimine PLP-GABA adduct | | Descriptor: | 4-AMINOBUTYRATE TRANSAMINASE, GAMMA-AMINO-BUTANOIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Bruce, H, Tuan, A.N, Mangas Sanchez, J, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-05-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of a Gamma-Aminobutyrate (Gaba) Transaminase from the S-Triazine-Degrading Organism Arthrobacter Aurescens Tc1 in Complex with Plp and with its External Aldimine Plp- Gaba Adduct.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
5G6S
 
 | | Imine reductase from Aspergillus oryzae in complex with NADP(H) and (R)-rasagiline | | Descriptor: | IMINE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RASAGILINE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-06-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A reductive aminase from Aspergillus oryzae.
Nat Chem, 9, 2017
|
|
6IAU
 
 | | Amine Dehydrogenase from Cystobacter fuscus in complex with NADP+ and cyclohexylamine | | Descriptor: | Amine Dehydrogenase, CYCLOHEXYLAMMONIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Beloti, L, Mayol, O, Turkenburg, J.P, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
4ATP
 
 | | Structure of GABA-transaminase A1R958 from Arthrobacter aurescens in complex with PLP | | Descriptor: | 4-AMINOBUTYRATE TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Bruce, H, Tuan, A.N, Mangas Sanchez, J, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-05-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a Gamma-Aminobutyrate (Gaba) Transaminase from the S-Triazine-Degrading Organism Arthrobacter Aurescens Tc1 in Complex with Plp and with its External Aldimine Plp- Gaba Adduct.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4BMV
 
 | | Short-chain dehydrogenase from Sphingobium yanoikuyae in complex with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SHORT-CHAIN DEHYDROGENASE | | Authors: | Man, H, Kedziora, K, Lavandera-Garcia, I, Gotor-Fernandez, V, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
5FJT
 
 | |
4D3S
 
 | | Imine reductase from Nocardiopsis halophila | | Descriptor: | IMINE REDUCTASE, octyl beta-D-glucopyranoside | | Authors: | Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, Activity and Stereoselectivity of Nadph-Dependent Oxidoreductases Catalysing the S-Selective Reduction of the Imine Substrate 2-Methylpyrroline.
Chembiochem, 16, 2015
|
|
5FJU
 
 | |
5FJR
 
 | |
5FJO
 
 | |
5FJP
 
 | |
