7LTA
 
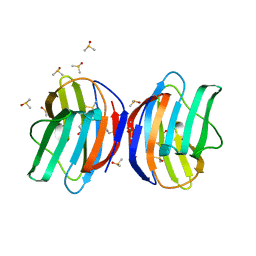 | | Galectin-1 in complex with Trehalose | | Descriptor: | DIMETHYL SULFOXIDE, Galectin-1, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Grimm, C, Bechold, J, Seibel, J. | | Deposit date: | 2021-02-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-1 in complex with Trehalose
To Be Published
|
|
4V98
 
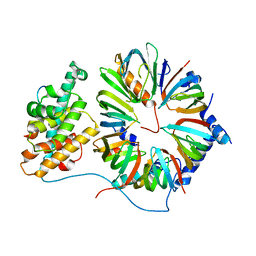 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
6EK5
 
 | |
5C8B
 
 | |
8P0J
 
 | |
8P0N
 
 | |
8P0K
 
 | |
6FME
 
 | |
4WKG
 
 | |
3H6Z
 
 | |
8B0Z
 
 | |
4FL9
 
 | |
3PTH
 
 | |
8B0W
 
 | |
5MAN
 
 | |
5MB2
 
 | |
5M9X
 
 | | Structure of sucrose phosphorylase from Bifidobacterium adolescentis bound to glycosylated resveratrol | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[3-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-5-oxidanyl-phenoxy]oxane-3,4 ,5-triol, Sucrose phosphorylase | | Authors: | Grimm, C, Kraus, M. | | Deposit date: | 2016-11-02 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Switching enzyme specificity from phosphate to resveratrol glucosylation.
Chem. Commun. (Camb.), 53, 2017
|
|
5MWT
 
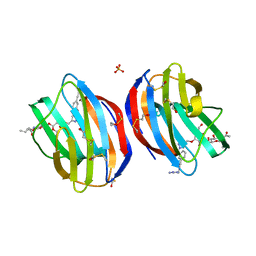 | | Galectin-1 in Complex with Ligand JB97 | | Descriptor: | BETA-MERCAPTOETHANOL, Galectin-1, SULFATE ION, ... | | Authors: | Grimm, C, Bechold, J. | | Deposit date: | 2017-01-19 | | Release date: | 2018-02-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural insights into the redesign of a sucrose phosphorylase by induced loop repositioning
To Be Published
|
|
5MWX
 
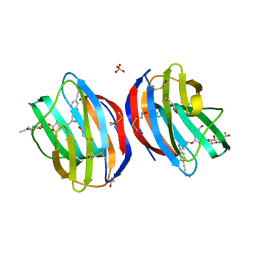 | | Galectin-1 in Complex with Ligand JB60 | | Descriptor: | BETA-MERCAPTOETHANOL, Galectin-1, SULFATE ION, ... | | Authors: | Grimm, C, Bechold, J. | | Deposit date: | 2017-01-20 | | Release date: | 2018-02-28 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Galectin-1 in Complex with Ligand JB60
To Be Published
|
|
6RIC
 
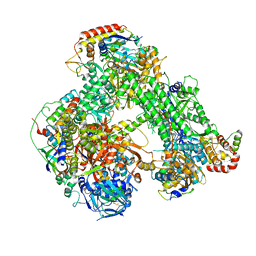 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
1FJH
 
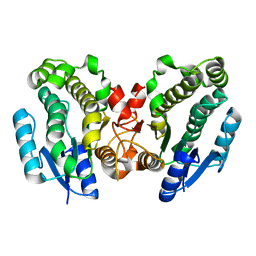 | | THE CRYSTAL STRUCTURE OF 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE FROM COMAMONAS TESTOSTERONI, A MEMBER OF THE SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY | | Descriptor: | 3ALPHA-HYDROXYSTEROID DEHYDROGENASE/CARBONYL REDUCTASE | | Authors: | Grimm, C, Ficner, R, Maser, E, Klebe, G, Reuter, K. | | Deposit date: | 2000-08-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of 3alpha -hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni shows a novel oligomerization pattern within the short chain dehydrogenase/reductase family.
J.Biol.Chem., 275, 2000
|
|
7AMV
 
 | | Atomic structure of the poxvirus transcription pre-initiation complex in the initially melted state | | Descriptor: | ATP-dependent helicase VETFS, DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOF
 
 | | Atomic structure of the poxvirus transcription late pre-initiation complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP8
 
 | | Atomic structure of the poxvirus initially transcribing complex in conformation 2 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOZ
 
 | | Atomic structure of the poxvirus transcription initiation complex in conformation 1 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
