1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
1HT3
 
 | |
1B1U
 
 | |
4LIZ
 
 | | Crystal structure of coactosin from Entamoeba histolytica | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Actin-binding protein, cofilin/tropomyosin family protein, ... | | Authors: | Gourinath, S, Kumar, N. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | EhCoactosin stabilizes actin filaments in the protist parasite Entamoeba histolytica.
Plos Pathog., 10, 2014
|
|
1FE5
 
 | | SEQUENCE AND CRYSTAL STRUCTURE OF A BASIC PHOSPHOLIPASE A2 FROM COMMON KRAIT (BUNGARUS CAERULEUS) AT 2.4 RESOLUTION: IDENTIFICATION AND CHARACTERIZATION OF ITS PHARMACOLOGICAL SITES. | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-01-24 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sequence and crystal structure determination of a basic phospholipase A2 from common krait (Bungarus caeruleus) at 2.4 A resolution: identification and characterization of its pharmacological sites.
J.Mol.Biol., 307, 2001
|
|
3BM5
 
 | | Crystal structure of O-acetyl-serine sulfhydrylase from Entamoeba histolytica in complex with cysteine | | Descriptor: | CYSTEINE, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Krishna, C, Kumar, M, Kumar, S, Gourinath, S. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase.
Proteins, 72, 2008
|
|
5FXT
 
 | |
8IW2
 
 | |
5JIS
 
 | |
5JJC
 
 | |
6KR5
 
 | |
5F8V
 
 | |
5XCH
 
 | | Crystal structure of Wild type Vps29 complexed with Zn+2 from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
5XCE
 
 | | Crystal structure of Wild type Vps29 from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
5XCK
 
 | | Crystal structure of Vps29 double mutant (D62A/H86A) from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
4NJM
 
 | |
4NFY
 
 | |
6M1X
 
 | |
8HRV
 
 | | dutpase of helicobacter pylori 26695 | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Kumari, K, Gourinath, S. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | dutpase of helicobacter pylori 26695
To Be Published
|
|
7VLJ
 
 | | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ali, M.F, Devi, S, Gourinath, S. | | Deposit date: | 2021-10-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation
To Be Published
|
|
1G2X
 
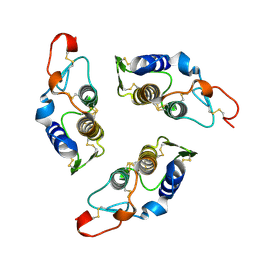 | | Sequence induced trimerization of krait PLA2: crystal structure of the trimeric form of krait PLA2 | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Paramsivam, M, Singh, T.P. | | Deposit date: | 2000-10-22 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-induced trimerization of phospholipase A2: structure of a trimeric isoform of PLA2 from common krait (Bungarus caeruleus) at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5JZX
 
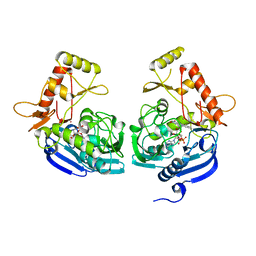 | | Crystal Structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Mycobacterium tuberculosis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Dharavath, S, Eniyan, K, Bajpai, U, Gourinath, S. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1866, 2017
|
|
1G0Z
 
 | |
5FRQ
 
 | |
3ULG
 
 | |
