3RFU
 
 | | Crystal structure of a copper-transporting PIB-type ATPase | | Descriptor: | Copper efflux ATPase, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Gourdon, P, Liu, X, Skjorringe, T, Morth, J.P, Birk Moller, L, Panyella Pedersen, B, Nissen, P. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a copper-transporting PIB-type ATPase.
Nature, 475, 2011
|
|
2RDD
 
 | | X-ray crystal structure of AcrB in complex with a novel transmembrane helix. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Acriflavine resistance protein B, UPF0092 membrane protein yajC | | Authors: | Tornroth-Horsefield, S, Gourdon, P, Horsefield, R, Neutze, R. | | Deposit date: | 2007-09-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist.
Structure, 15, 2007
|
|
8QKK
 
 | | Cryo-EM structure of MmpL3 from Mycobacterium smegmatis reconstituted into peptidiscs | | Descriptor: | Trehalose monomycolate exporter MmpL3 | | Authors: | Couston, J, Guo, Z, Wang, K, Gourdon, P.E, Blaise, M. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the trehalose monomycolate transporter, MmpL3, reconstituted into peptidiscs.
Curr Res Struct Biol, 6, 2023
|
|
4UMV
 
 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN THE E2P STATE | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
7NNH
 
 | |
4UMW
 
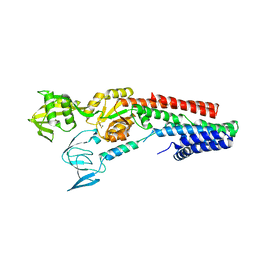 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN E2.PI STATE | | Descriptor: | MAGNESIUM ION, TETRAFLUOROALUMINATE ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
8C9H
 
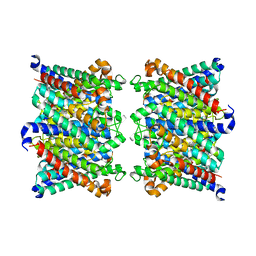 | | AQP7_inhibitor | | Descriptor: | Aquaporin-7, ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate | | Authors: | Huang, P, Venskutonyte, R, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2023-01-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for human aquaporin inhibition.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6F7H
 
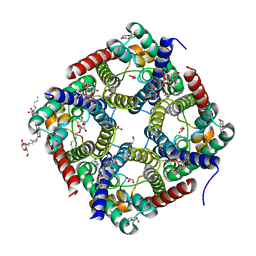 | | Crystal structure of human AQP10 | | Descriptor: | Aquaporin-10, GLYCEROL, nonyl beta-D-glucopyranoside | | Authors: | Gotfryd, K, Wang, K, Missel, J.W, Pedersen, P.A, Gourdon, P. | | Deposit date: | 2017-12-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Human adipose glycerol flux is regulated by a pH gate in AQP10.
Nat Commun, 9, 2018
|
|
5I32
 
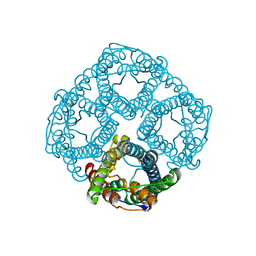 | | Ammonia permeable aquaporin AtTIP2;1 | | Descriptor: | Aquaporin TIP2-1 | | Authors: | Kirscht, A, Nissen, P, Kjellbom, P, Gourdon, P, Johanson, U. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of an Ammonia-Permeable Aquaporin.
Plos Biol., 14, 2016
|
|
4HQJ
 
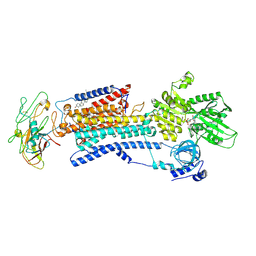 | | Crystal structure of Na+,K+-ATPase in the Na+-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Nyblom, M, Reinhard, L, Gourdon, P, Nissen, P. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-02 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state.
Science, 342, 2013
|
|
7Z6N
 
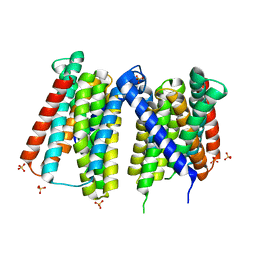 | | Crystal structure of Zn2+-transporter BbZIP in a metal-stripped state | | Descriptor: | Putative membrane protein, SULFATE ION | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
7Z6M
 
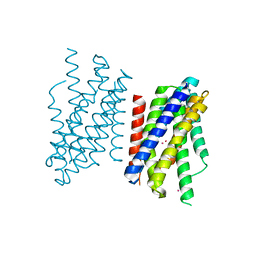 | | Crystal structure of Zn2+-transporter BbZIP in a cadmium bound state | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
7B52
 
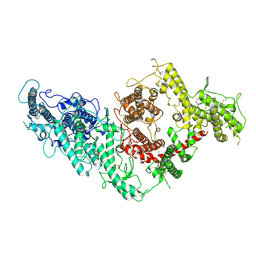 | | VAR2CSA full ectodomain | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Gourdon, P.E, Dagil, R, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
7B54
 
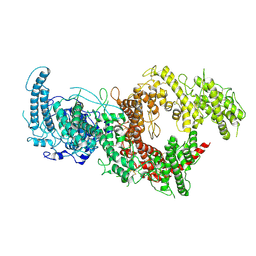 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
7PRU
 
 | |
7PSD
 
 | |
7PRO
 
 | |
7PQX
 
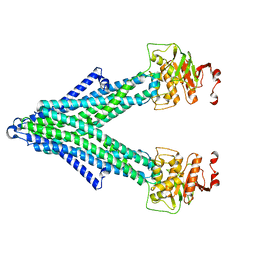 | |
7PR1
 
 | |
7QBZ
 
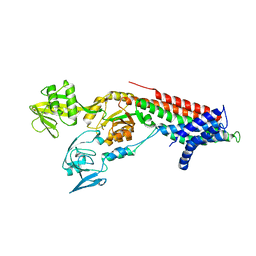 | | Crystal structure Cadmium translocating P-type ATPase | | Descriptor: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QC0
 
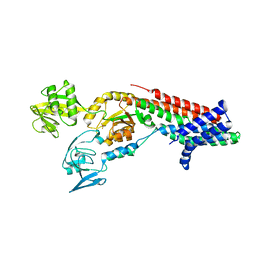 | | Crystal structure of Cadmium translocating P-type ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7R0I
 
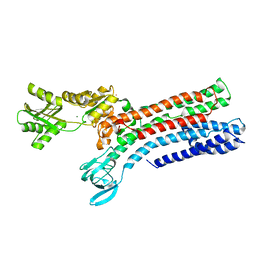 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0H
 
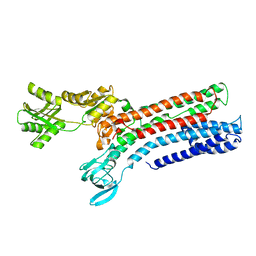 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | COPPER (II) ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0G
 
 | |
7OP3
 
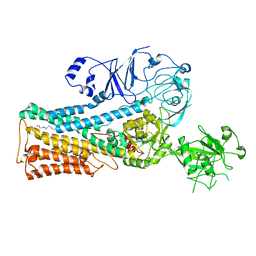 | | Cryo-EM structure of P5B-ATPase E2PiSPM | | Descriptor: | Cation-transporting ATPase, SPERMINE | | Authors: | Li, P, Gronberg, C, Wang, K.T, Salustros, N, Gourdon, P.E. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
