8PWR
 
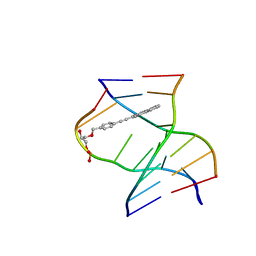 | | TINA-conjugated antiparallel DNA triplex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | Authors: | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | Deposit date: | 2023-07-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
2BNK
 
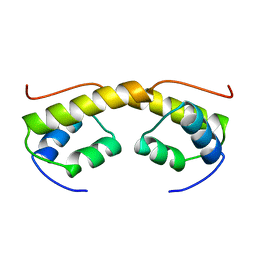 | | The structure of phage phi29 replication organizer protein p16.7 | | Descriptor: | EARLY PROTEIN GP16.7 | | Authors: | Albert, A, Asensio, J.L, Munoz-Espin, D, Gonzalez, C, Hermoso, J.A, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-03-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Functional Domain of {Varphi}29 Replication Organizer: Insights Into Oligomerization and DNA Binding.
J.Biol.Chem., 280, 2005
|
|
2C3E
 
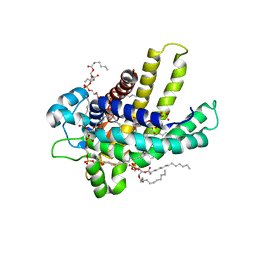 | | The bovine mitochondrial ADP-ATP carrier | | Descriptor: | ADP/ATP TRANSLOCASE 1, CARDIOLIPIN, Carboxyatractyloside | | Authors: | Nury, H, Dahout-Gonzalez, C, Trezeguet, V, Lauquin, G, Brandolin, G, Pebay-Peyroula, E. | | Deposit date: | 2005-10-06 | | Release date: | 2005-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Lipid-Mediated Interactions between Mitochondrial Adp/ATP Carrier Monomers.
FEBS Lett., 579, 2005
|
|
2KSS
 
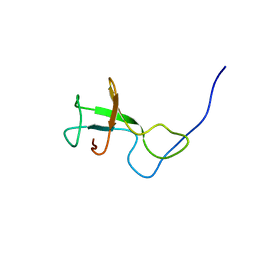 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
2HK4
 
 | |
6FC9
 
 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
1ZAE
 
 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1GPT
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-H THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1IEK
 
 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER S) | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-07-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1IEY
 
 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER R) | | Descriptor: | 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', 5'-D(P*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-11 | | Release date: | 2001-07-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1J9N
 
 | | Solution Structure of the Nucleopeptide [AC-LYS-TRP-LYS-HSE(p3*dGCATCG)-ALA]-[p5*dCGTAGC] | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*CP*(PGN))-3', peptide ACE-LYS-TRP-LYS-HSE-ALA | | Authors: | Gomez-Pinto, I, Marchan, V, Gago, F, Grandas, A, Gonzalez, C. | | Deposit date: | 2001-05-28 | | Release date: | 2003-01-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of tryptophan-containing nucleopeptide duplexes
Chembiochem, 4, 2003
|
|
8BV6
 
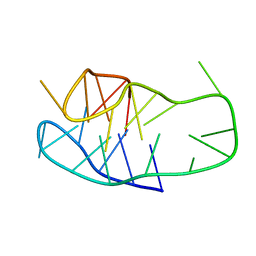 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BQY
 
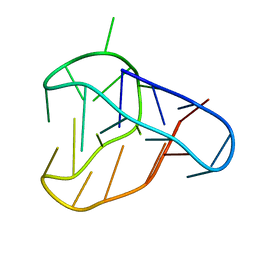 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
1OKC
 
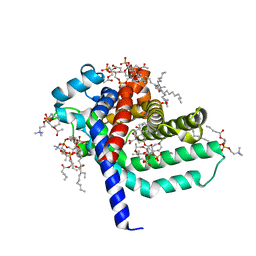 | | structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-LAURYLAMIDO-N,N'-DIMETHYLPROPYLAMINOXYDE, ADP, ... | | Authors: | Pebay-Peyroula, E, Dahout-Gonzalez, C, Kahn, R, Trezeguet, V, Lauquin, G.J.-M, Brandolin, G. | | Deposit date: | 2003-07-21 | | Release date: | 2003-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mitochondrial Adp/ATP Carrier in Complex with Carboxyatractyloside
Nature, 426, 2003
|
|
1PNB
 
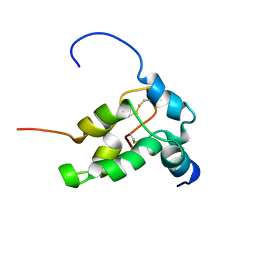 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | Descriptor: | NAPIN BNIB | | Authors: | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | Deposit date: | 1996-09-17 | | Release date: | 1997-09-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
1SP6
 
 | | A DNA duplex containing a cholesterol adduct (alpha-anomer) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOL)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
1SSJ
 
 | | A DNA DUPLEX CONTAINING A CHOLESTEROL ADDUCT (BETA-ANOMER) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOB)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
2LSC
 
 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
7OGV
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*(P*GP*CP*GP*TP*(DH)P*GP*AP*CP*GP*CP*G-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-11-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7O5E
 
 | |
7OHM
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*(DH)P*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHJ
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHE
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*CP*GP*AP*CP*GP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
