5TSR
 
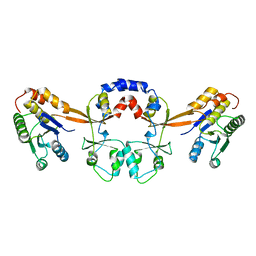 | |
5UM3
 
 | |
5V46
 
 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | Descriptor: | Sacsin | | Authors: | Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V45
 
 | |
5V47
 
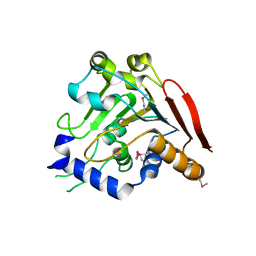
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5VSZ
 
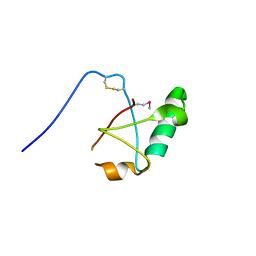 | |
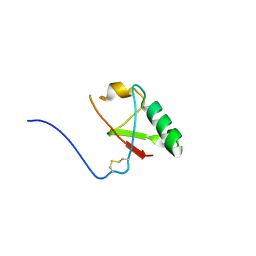
5VSX
 
 | |
1FC8
 
 | |
1GH8
 
 | |
1IFW
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1JDQ
 
 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCU
 
 | |
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
