8J5J
 
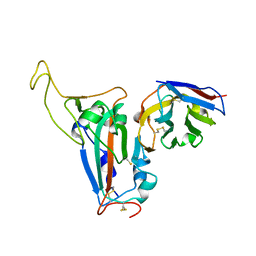 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
4K65
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
6AEE
 
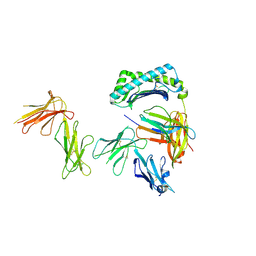 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
4F7P
 
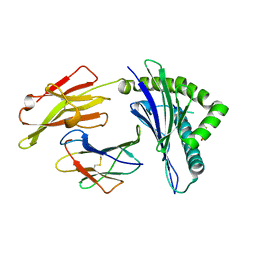 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4MJ5
 
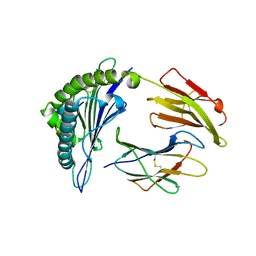 | | Crystal Structure of HLA-A*1101 in complex with H1-22, an influenza A(H1N1) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
4MJ6
 
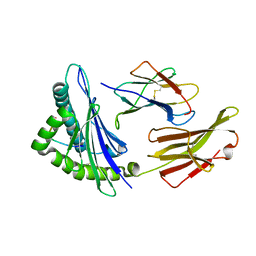 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
8I5C
 
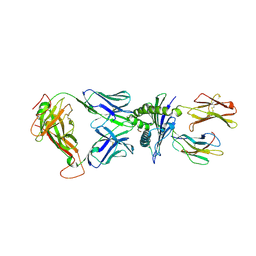 | | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), TCR alpha chain, ... | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-24 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
6ISC
 
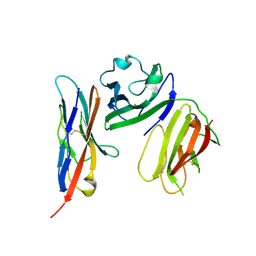 | | complex structure of mCD226-ecto and hCD155-D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISA
 
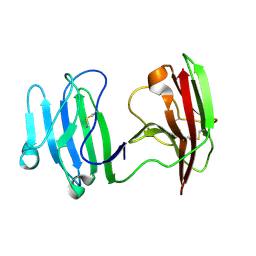 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISB
 
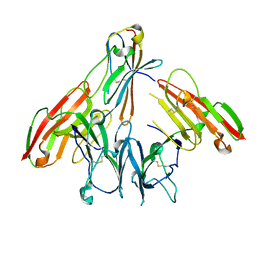 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J5G
 
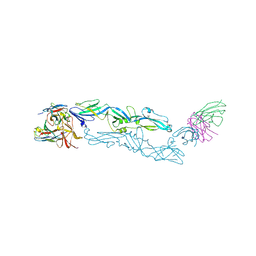 | | Complex structure of MAb 4.2-scFv with tick-borne encephalitis virus envelope protein | | Descriptor: | Envelope protein E, antibody heavy chain, antibody light chain | | Authors: | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.291 Å) | | Cite: | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
6J5C
 
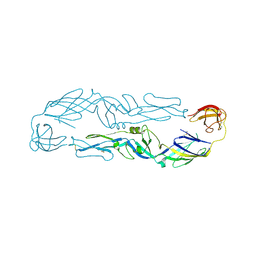 | | Louping ill virus envelope protein | | Descriptor: | Envelope protein E | | Authors: | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
6J5D
 
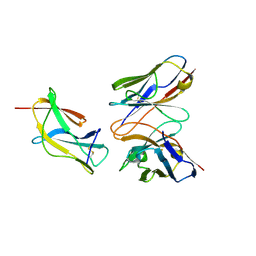 | | Complex structure of MAb 4.2-scFv with louping ill virus envelope protein Domain III | | Descriptor: | Envelope, antibody heavy chain, antibody light chain | | Authors: | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
6J5F
 
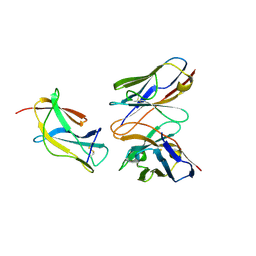 | | Complex structure of MAb 4.2-scFv with tick-borne encephalitis virus envelope protein Domain III | | Descriptor: | Envelope protein, antibody heavy chain, antibody light chain | | Authors: | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
6J2D
 
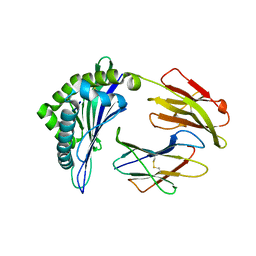 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | HeV1, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
8KC2
 
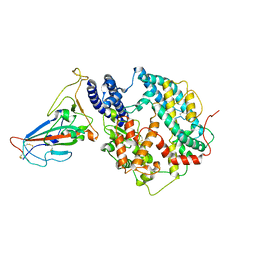 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
6KLE
 
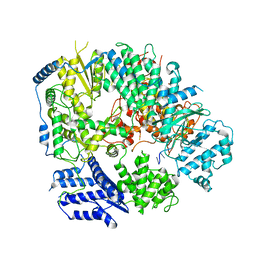 | | Monomeric structure of Machupo virus polymerase bound to vRNA promoter | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
4HX1
 
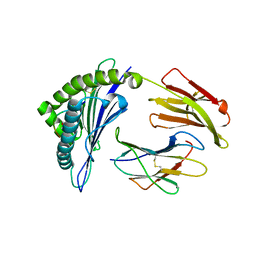 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
5XLP
 
 | |
4HZW
 
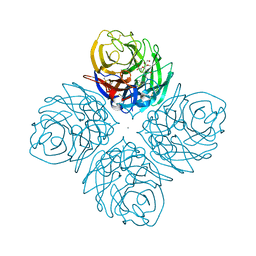 | | Crystal structure of influenza A neuraminidase N3 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZX
 
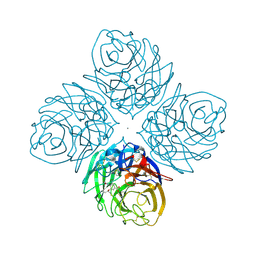 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
5XJ4
 
 | | Complex structure of durvalumab-scFv/PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab-VH, durvalumab-VL | | Authors: | Tan, S, Liu, K, Chai, Y, Gao, G.F, Qi, J. | | Deposit date: | 2017-04-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab
Protein Cell, 9, 2018
|
|
4HZZ
 
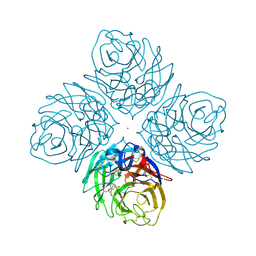 | | Crystal structure of influenza neuraminidase N3-H274Y complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZV
 
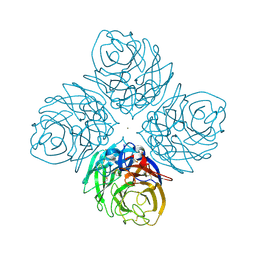 | | The crystal structure of influenza A neuraminidase N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
