3HCJ
 
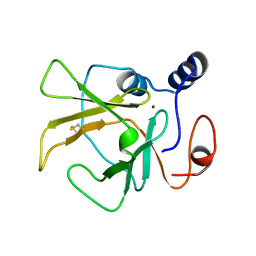 | |
2J89
 
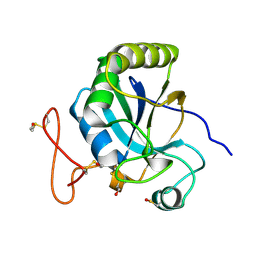 | | Functional and structural aspects of poplar cytosolic and plastidial type A methionine sulfoxide reductases | | Descriptor: | BETA-MERCAPTOETHANOL, METHIONINE SULFOXIDE REDUCTASE A | | Authors: | Rouhier, N, Kauffmann, B, Tete-Favier, F, Palladino, P, Gans, P, Branlant, G, Jacquot, J.P, Boschi-Muller, S. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Aspects of Poplar Cytosolic and Plastidial Type a Methionine Sulfoxide Reductases
J.Biol.Chem., 282, 2007
|
|
3BQG
 
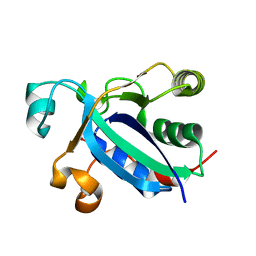 | |
3BQE
 
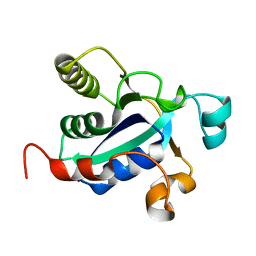 | |
3BQH
 
 | |
3BQF
 
 | |
4F03
 
 | |
4F0B
 
 | |
4F0C
 
 | | Crystal structure of the glutathione transferase URE2P5 from Phanerochaete chrysosporium | | Descriptor: | GLYCEROL, Glutathione transferase, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Didierjean, C, Favier, F, Roret, T. | | Deposit date: | 2012-05-04 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary divergence of Ure2pA glutathione transferases in wood degrading fungi.
Fungal Genet Biol, 83, 2015
|
|
4G19
 
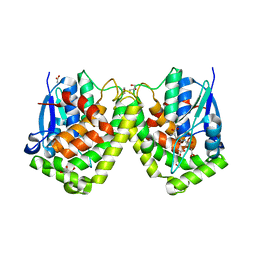 | | Crystal structure of the glutathione transferase GTE1 from Phanerochaete chrysosporium in complex with glutathione | | Descriptor: | ACETATE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Didierjean, C, Favier, F, Prosper, P. | | Deposit date: | 2012-07-10 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Phanerochaete chrysosporium Glutathione Transferase Reveals a Novel Structural and Functional Class with Ligandin Properties.
J.Biol.Chem., 287, 2012
|
|
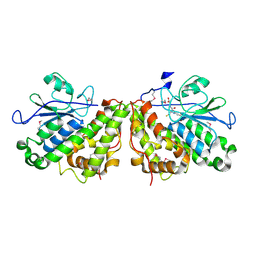
3PPU
 
 | |
4G10
 
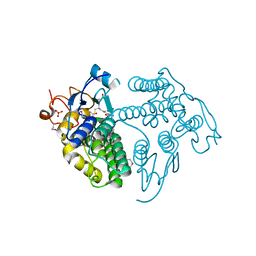 | | LigG from Sphingobium sp. SYK-6 is related to the glutathione transferase omega class | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase homolog, ... | | Authors: | Meux, E, Prosper, P, Masai, E, Mulliert Carlin, G, Dumarcay, S, Morel, M, Didierjean, C, Gelhaye, E, Favier, F. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class.
Febs Lett., 586, 2012
|
|
6GIC
 
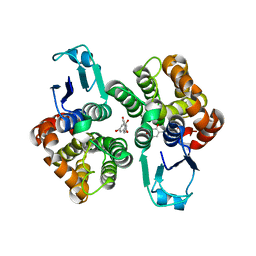 | |
6HT6
 
 | | Crystal structure of glutathione transferase Omega 2S from Trametes versicolor in complex with 2,4-dihydroxybenzophenone | | Descriptor: | Glutathione S-transferase omega 2S, MAGNESIUM ION, [2,4-bis(oxidanyl)phenyl]-phenyl-methanone | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6GCB
 
 | |
6GCC
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
6GIB
 
 | |
6HPE
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with the glutathione adduct of phenethyl-isothiocyanate | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of Trametes versicolor glutathione transferase Omega 3S bound to its conjugation product glutathionyl-phenethylthiocarbamate reveals plasticity of its active site.
Protein Sci., 28, 2019
|
|
6HTA
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Glutathione transferase Xi 3 C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
1NQO
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQ5
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQA
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
6GC9
 
 | |
6GCA
 
 | |
