6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6CZG
 
 | |
6CZJ
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
8ETQ
 
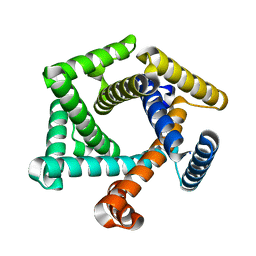 | |
7R9G
 
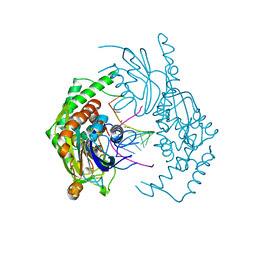 | | Catalytically inactive yeast Pseudouridine Synthase, PUS1, bound to RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*AP*AP*UP*CP*GP*GP*GP*AP*UP*UP*CP*CP*GP*GP*AP*UP*A)-3'), SULFATE ION, ... | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1.
Plos One, 18, 2023
|
|
7R9F
 
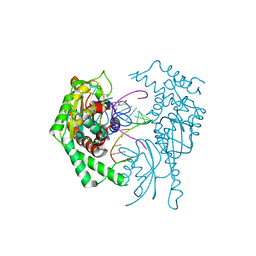 | |
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
5BVB
 
 | | Engineered Digoxigenin binder DIG5.1a | | Descriptor: | DIG5.1a, DIGOXIGENIN | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2015-06-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge.
J.Chem.Inf.Model., 56, 2016
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
3B5L
 
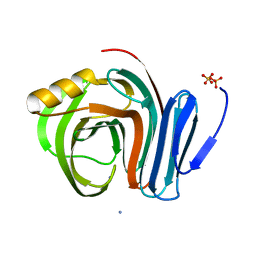 | |
3HOJ
 
 | |
4J8T
 
 | | Engineered Digoxigenin binder DIG10.2 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.2 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4J9A
 
 | | Engineered Digoxigenin binder DIG10.3 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.3 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
5IEO
 
 | | Structure of CDL2.3a, a computationally designed Vitamin-D3 binder | | Descriptor: | 1,2-ETHANEDIOL, 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3a | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IER
 
 | |
5IEP
 
 | | Structure of CDL2.3b, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3b | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IF6
 
 | |
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
