7BH2
 
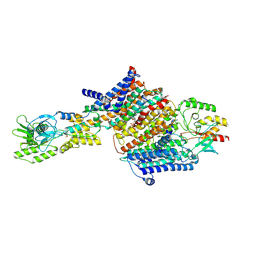 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3KG7
 
 | |
7BH1
 
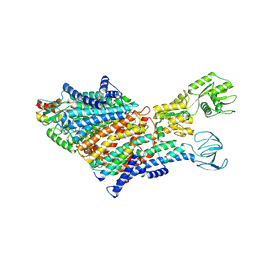 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BGY
 
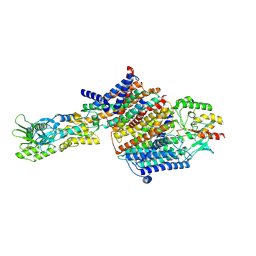 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3KQ0
 
 | | Crystal structure of human alpha1-acid glycoprotein | | Descriptor: | (2R)-2,3-dihydroxypropyl acetate, Alpha-1-acid glycoprotein 1, CHLORIDE ION | | Authors: | Schiefner, A, Schonfeld, D.L, Ravelli, R.B.G, Mueller, U, Skerra, A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin.
J.Mol.Biol., 384, 2008
|
|
3LCR
 
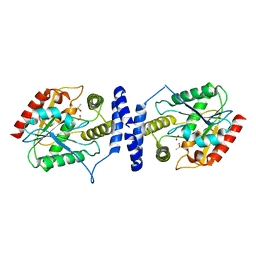 | | Thioesterase from Tautomycetin Biosynthhetic Pathway | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Tautomycetin biosynthetic PKS | | Authors: | Akey, D.L, Scaglione, J.B, Smith, J.L, Sherman, D.H. | | Deposit date: | 2010-01-11 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3MJ4
 
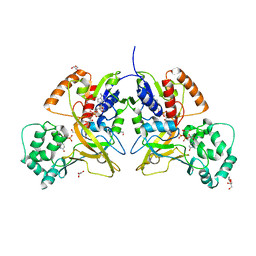 | | Crystal structure of UDP-galactopyranose mutase in complex with phosphonate analog of UDP-galactopyranose | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Karunan Partha, S, Sadeghi-Khomami, A, Slowski, K, Kotake, T, Thomas, N.R, Jakeman, D.L, Sanders, D.A.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemoenzymatic Synthesis, Inhibition Studies, and X-ray Crystallographic Analysis of the Phosphono Analog of UDP-Galp as an Inhibitor and Mechanistic Probe for UDP-Galactopyranose Mutase.
J.Mol.Biol., 403, 2010
|
|
6O0E
 
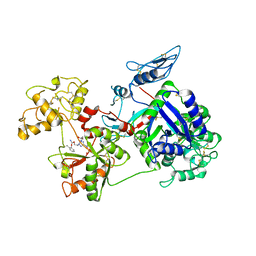 | | Saxiphilin:STX complex, soaking | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6O2U
 
 | | Crystal structure of the SARAF luminal domain | | Descriptor: | Store-operated calcium entry-associated regulatory factor | | Authors: | Kimberlin, C.R, Minor, D.L. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARAF Luminal Domain Structure Reveals a Novel Domain-Swapped beta-Sandwich Fold Important for SOCE Modulation.
J.Mol.Biol., 431, 2019
|
|
6O0D
 
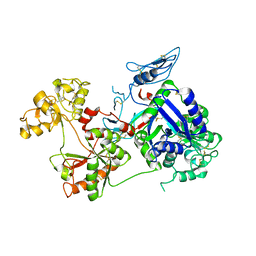 | | Saxiphilin Apo structure | | Descriptor: | Saxiphilin | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6O2W
 
 | |
6O0F
 
 | | Saxiphilin:STX complex, co-crystal | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6O2V
 
 | |
3NM9
 
 | | HMGD(M13A)-DNA complex | | Descriptor: | DNA 5'-D(*G*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3', High mobility group protein D | | Authors: | Churchill, M.E.A, Klass, J, Zoetewey, D.L. | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending.
J.Mol.Biol., 403, 2010
|
|
3NA7
 
 | | 2.2 Angstrom Structure of the HP0958 Protein from Helicobacter pylori CCUG 17874 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HP0958, MAGNESIUM ION, ... | | Authors: | Caly, D.L, O'Toole, P.W, Moore, S.A. | | Deposit date: | 2010-06-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2-A Structure of the HP0958 Protein from Helicobacter pylori Reveals a Kinked Anti-Parallel Coiled-Coil Hairpin Domain and a Highly Conserved Zn-Ribbon Domain
J.Mol.Biol., 403, 2010
|
|
3O4O
 
 | | Crystal structure of an Interleukin-1 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, ... | | Authors: | Wang, X.Q, Wang, D.L, Zhang, S.Y, Li, L, Liu, X, Mei, K.R. | | Deposit date: | 2010-07-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the assembly and activation of IL-1beta with its receptors
Nat.Immunol., 11, 2010
|
|
2OVC
 
 | |
2P4F
 
 | |
2P4X
 
 | |
8D00
 
 | |
8E77
 
 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E75
 
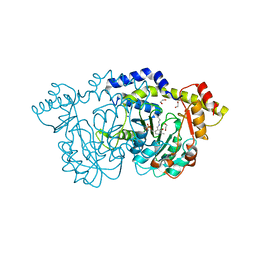 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E62
 
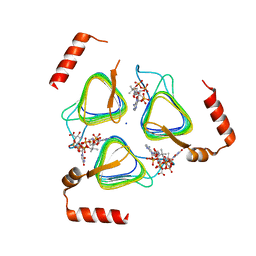 | | STRUCTURE OF Pcryo_0615 from Psychrobacter cryohalolentis, an N-acetyltransferase required to produce Diacetamido-2,3-dideoxy-D-glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, COENZYME A, SODIUM ION, ... | | Authors: | Hofmeister, D.L, Bockhaus, N.J, Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
2Q99
 
 | |
2QFG
 
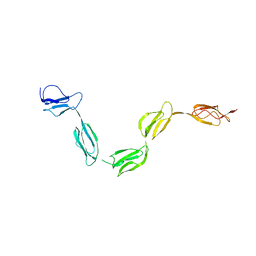 | | Solution Structure of the N-terminal SCR-1/5 fragment of Complement Factor H. | | Descriptor: | Complement factor H | | Authors: | Okemefuna, A.I, Gilbert, H.E, Griggs, K.M, Ormsby, R.J, Gordon, D.L, Perkins, S.J. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties.
J.Mol.Biol., 375, 2008
|
|
