1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOJ
 
 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
6P7P
 
 | | Structure of E. coli MS115-1 NucC, cAAA-bound form | | Descriptor: | CHLORIDE ION, Cyclic tri-AMP (5'-3' linked), E. coli MS115-1 NucC 2-241 | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P80
 
 | | Structure of E. coli MS115-1 CdnC + ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Azimi, C.S, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8P
 
 | | Structure of P. aeruginosa ATCC27853 HORMA1 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8R
 
 | | Structure of P. aeruginosa ATCC27853 HORMA2 | | Descriptor: | HORMA domain containing protein, PHOSPHATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6PB3
 
 | | Structure of Rhizobiales Trip13 | | Descriptor: | Rhizobiales Sp. Pch2, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P82
 
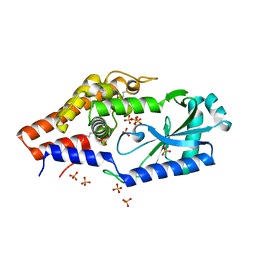 | | Structure of P. aeruginosa ATCC27853 CdnD, Apo form 1 | | Descriptor: | Nucleotidyltransferase, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P7O
 
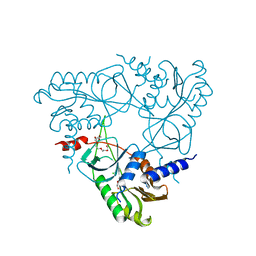 | | Structure of E. coli MS115-1 NucC, Apo form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, HEXAETHYLENE GLYCOL, ... | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P7Q
 
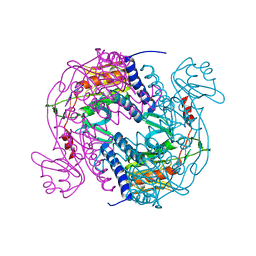 | | Structure of E. coli MS115-1 NucC, 5'-pApA bound form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, RNA (5'-R(P*AP*A)-3') | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8J
 
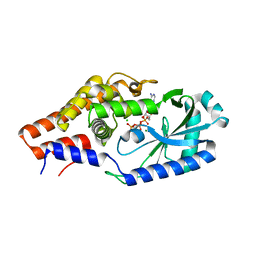 | |
6P8S
 
 | |
6P8U
 
 | | Structure of P. aeruginosa ATCC27853 CdnD:HORMA2:Peptide 1 complex | | Descriptor: | HORMA domain containing protein, MAGNESIUM ION, Nucleotidyltransferase, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6Q1H
 
 | |
7SQR
 
 | | 201phi2-1 Chimallin localized tetramer reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQQ
 
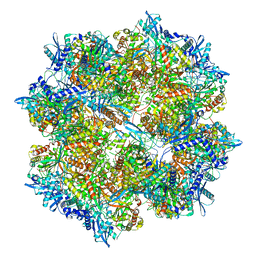 | | 201Phi2-1 Chimallin Cubic (O, 24mer) assembly | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQU
 
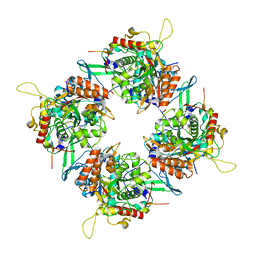 | | Goslar chimallin C4 tetramer localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQV
 
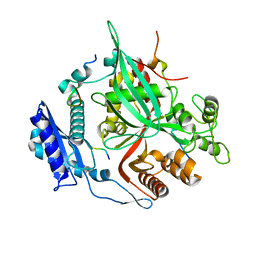 | | Goslar chimallin C1 localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQS
 
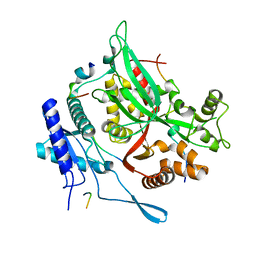 | | 201phi2-1 Chimallin C1 localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQT
 
 | | Goslar chimallin cubic (O, 24mer) assembly | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7T5U
 
 | | Structure of E. coli MS115-1 CapH N-terminal domain | | Descriptor: | Helix-turn-helix domain-containing protein, SULFATE ION | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
7T5V
 
 | |
7T5T
 
 | | Structure of Thauera sp. K11 CapP | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CapP toxin, SULFATE ION, ... | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
