7SR1
 
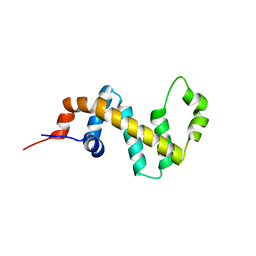 | |
7SR2
 
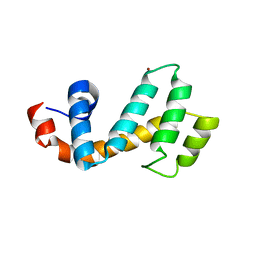 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
6N5Z
 
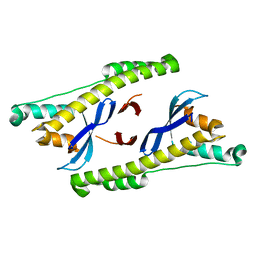 | |
6N5X
 
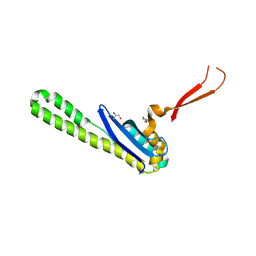 | | Crystal structure of the SNX5 PX domain in complex with the CI-MPR (space group P212121 - Form 1) | | Descriptor: | GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Sorting nexin-5,Cation-independent mannose-6-phosphate receptor | | Authors: | Collins, B, Paul, B, Weeratunga, S. | | Deposit date: | 2018-11-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins.
Nat.Cell Biol., 21, 2019
|
|
6N5Y
 
 | |
6MD5
 
 | |
4QDH
 
 | |
5TGH
 
 | |
1MGQ
 
 | |
4YMR
 
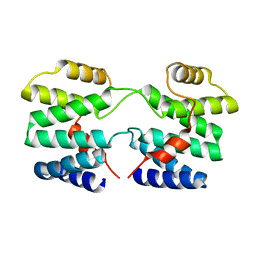 | |
1Z2W
 
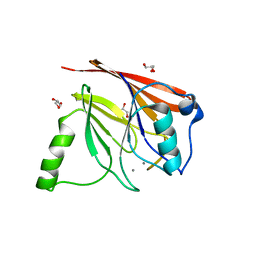 | | Crystal structure of mouse Vps29 complexed with Mn2+ | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
5W8M
 
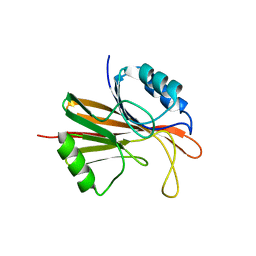 | | Crystal structure of Chaetomium thermophilum Vps29 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, Vacuolar protein sorting-associated protein 29 | | Authors: | Collins, B.M, Leneva, N. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
5UFC
 
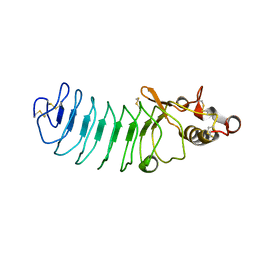 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 with H-trisaccharide bound | | Descriptor: | Tn4-22, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
1Z2X
 
 | | Crystal structure of mouse Vps29 | | Descriptor: | Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
5UFF
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 with Fucose(alpha-1-2)Lactose bound | | Descriptor: | RBC36, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFD
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 (Apo) | | Descriptor: | MAGNESIUM ION, RBC36 | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5WK4
 
 | | Crystal structure of an anti-idiotype VLR | | Descriptor: | MAGNESIUM ION, Variable lymphocyte receptor 39 | | Authors: | Collins, B.C, Nakahara, H, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of an anti-idiotype variable lymphocyte receptor.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5TGJ
 
 | |
4K60
 
 | | Crystal Structure of Human Chymase in Complex with Fragment 6-bromo-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromo-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4K2Y
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4P2J
 
 | |
4K69
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Linked Benzimidazolone Inhibitor: (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid | | Descriptor: | (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4K5Z
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
5TGI
 
 | |
1N9S
 
 | | Crystal structure of yeast SmF in spacegroup P43212 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
