1T26
 
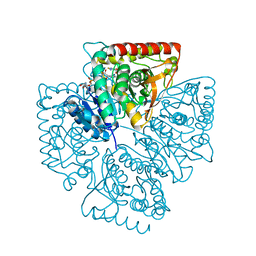 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-HYDROXY-1,2,5-THIADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1U5A
 
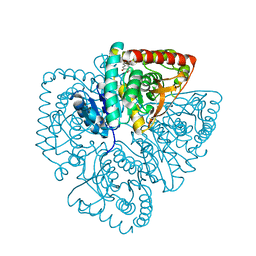 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,5-dihydroxy-2-naphthoic acid | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U4S
 
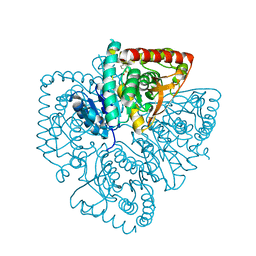 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedisulphonic acid | | Descriptor: | L-lactate dehydrogenase, NAPHTHALENE-2,6-DISULFONIC ACID | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U4O
 
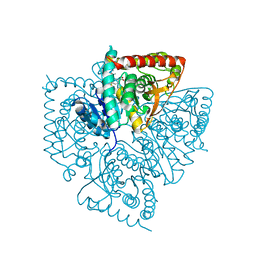 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedicarboxylic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,6-DICARBOXYNAPHTHALENE, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U5C
 
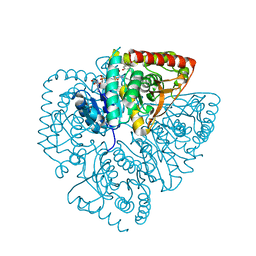 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
4CUJ
 
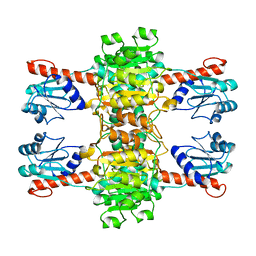 | |
4CUK
 
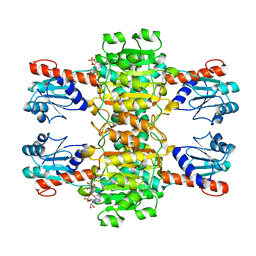 | |
4DZN
 
 | | A de novo designed Coiled Coil CC-pIL | | Descriptor: | COILED-COIL PEPTIDE CC-PIL | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZM
 
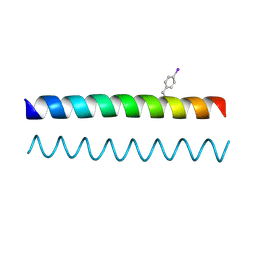 | | A de novo designed Coiled Coil CC-Di | | Descriptor: | COILED-COIL PEPTIDE CC-DI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZL
 
 | | A de novo designed Coiled Coil CC-Tri | | Descriptor: | COILED-COIL PEPTIDE CC-TRI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZK
 
 | | A de novo designed Coiled Coil CC-Tri-N13 | | Descriptor: | CHLORIDE ION, COILED-COIL PEPTIDE CC-TRI-N13 | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4H8F
 
 | |
4H8G
 
 | |
4H8L
 
 | |
4H8O
 
 | |
3NTN
 
 | | Crystal Structure of UspA1 head and neck domain from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Conners, R, Zaccai, N, Agnew, C, Burton, N, Brady, R.L. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1AE6
 
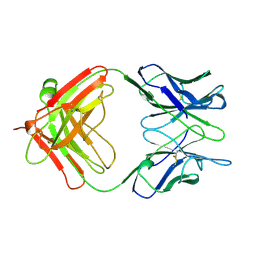 | | IGG-FAB FRAGMENT OF MOUSE MONOCLONAL ANTIBODY CTM01 | | Descriptor: | IGG CTM01 FAB (HEAVY CHAIN), IGG CTM01 FAB (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-03-06 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1AD0
 
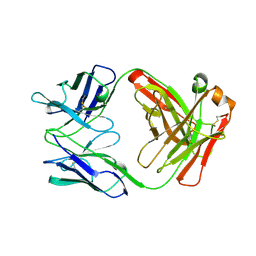 | | FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 | | Descriptor: | ANTIBODY A5B7 (HEAVY CHAIN), ANTIBODY A5B7 (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1AD9
 
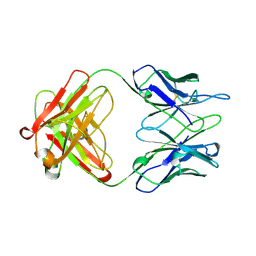 | | IGG-FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY CTM01 | | Descriptor: | IGG CTM01 FAB (HEAVY CHAIN), IGG CTM01 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-24 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1A7B
 
 | |
1A6P
 
 | |
1A64
 
 | |
1B96
 
 | |
1B97
 
 | |
1B94
 
 | | RESTRICTION ENDONUCLEASE ECORV WITH CALCIUM | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), RESTRICTION ENDONUCLEASE ECORV | | Authors: | Thomas, M.P, Halford, S.E, Brady, R.L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group.
Nucleic Acids Res., 27, 1999
|
|
