4NL8
 
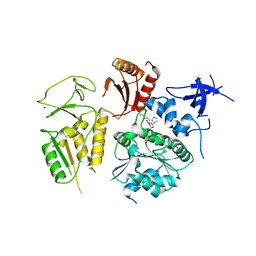 | | PriA Helicase Bound to SSB C-terminal Tail Peptide | | Descriptor: | Primosome assembly protein PriA, Single-stranded DNA-binding protein, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Thurmes, T.M, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.08 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NL4
 
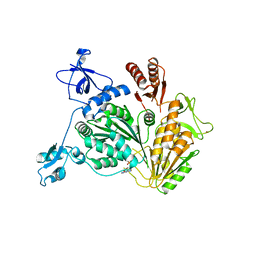 | | PriA Helicase Bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primosome assembly protein PriA, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5SZ8
 
 | |
6E8A
 
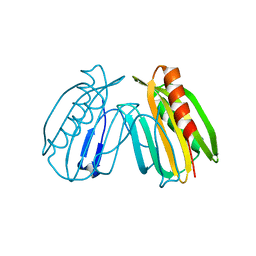 | | Crystal structure of DcrB from Salmonella enterica at 1.92 Angstroms resolution | | Descriptor: | DUF1795 domain-containing protein | | Authors: | Rasmussen, D.M, Soens, R.W, Bhattacharyya, B, May, J.F. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
J. Struct. Biol., 204, 2018
|
|
8SBS
 
 | |
6NZC
 
 | |
6NZA
 
 | |
6NZB
 
 | |
5KKD
 
 | |
6PZL
 
 | |
6PYK
 
 | |
5KDK
 
 | |
5KEH
 
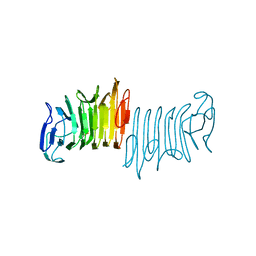 | |
5KF3
 
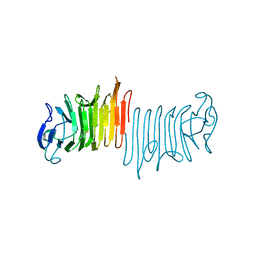 | |
