7UDN
 
 | |
7UPO
 
 | |
7UPP
 
 | |
7UE2
 
 | |
6V0F
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 4 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0H
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 6 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0C
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 1 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0E
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 3 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0J
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 8 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0G
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 5 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0I
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 7 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6VCI
 
 | | Lipophilic envelope-spanning tunnel protein (LetB), domains MCE2-MCE3 | | Descriptor: | Lipophilic envelope-spanning tunnel protein LetB | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-12-21 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0D
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 2 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
7MI8
 
 | | Signal subtracted reconstruction of AAA5 and AAA6 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 5 | | Descriptor: | Fusion protein of Dynein and Endolysin | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI6
 
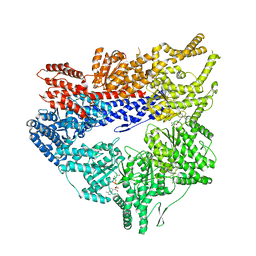 | | Yeast dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound, Model 1 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Urnavicius, L, Coudray, N, Ekeirt, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI3
 
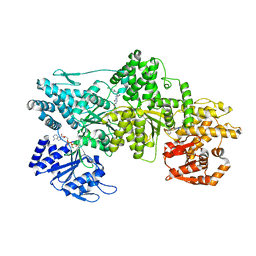 | | Signal subtracted reconstruction of AAA2, AAA3, and AAA4 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 4 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI1
 
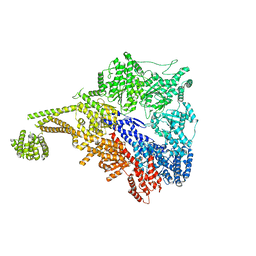 | |
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
4KJL
 
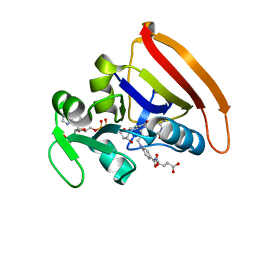 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
