5D1Y
 
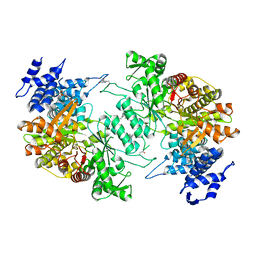 | | Low resolution crystal structure of human ribonucleotide reductase alpha6 hexamer in complex with dATP | | Descriptor: | Ribonucleoside-diphosphate reductase large subunit | | Authors: | Ando, N, Li, H, Brignole, E.J, Thompson, S, McLaughlin, M.I, Page, J, Asturias, F, Stubbe, J, Drennan, C.L. | | Deposit date: | 2015-08-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (9.005 Å) | | Cite: | Allosteric Inhibition of Human Ribonucleotide Reductase by dATP Entails the Stabilization of a Hexamer.
Biochemistry, 55, 2016
|
|
8DYZ
 
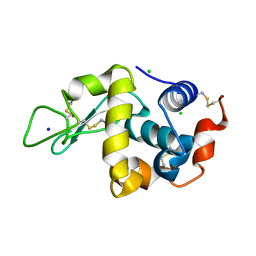 | |
8DZ7
 
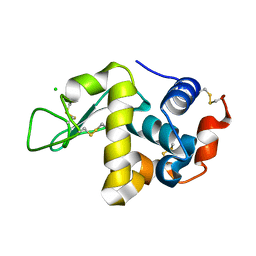 | |
6M8F
 
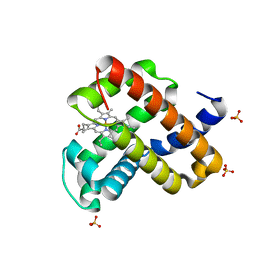 | | Engineered sperm whale myoglobin-based carbene transferase | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Bacik, J.P, Ando, N, Fasan, R. | | Deposit date: | 2018-08-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Origin of high stereocontrol in olefin cyclopropanation catalyzed by an engineered carbene transferase.
Acs Catalysis, 9, 2019
|
|
6UYK
 
 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
8ESU
 
 | | Myoglobin variant Mb-imi complex | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
8ESS
 
 | | Myoglobin variant Mb-cIII complex | | Descriptor: | Myoglobin, SULFATE ION, [2,18-bis(2-carboxyethyl)-7,12-diethenyl-3,8,13,17-tetramethyl-21-(2-oxo-3-phenylpropyl)porphyrin-21-iumato(2-)-kappa~3~N~22~,N~23~,N~24~]iron(2+) | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
7SPG
 
 | | Crystal structure of sperm whale myoglobin variant sMb13(pCaaF) in space group P212121 | | Descriptor: | CHLORIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tuning Enzyme Thermostability via Computationally Guided Covalent Stapling and Structural Basis of Enhanced Stabilization.
Biochemistry, 61, 2022
|
|
7SPH
 
 | | Crystal structure of sperm whale myoglobin variant sMb13(pCaaF) in space group P21 | | Descriptor: | Myoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tuning Enzyme Thermostability via Computationally Guided Covalent Stapling and Structural Basis of Enhanced Stabilization.
Biochemistry, 61, 2022
|
|
7SPE
 
 | |
7SPF
 
 | | Crystal structure of sperm whale myoglobin variant sMb5(pCaaF) | | Descriptor: | Myoglobin, NITRATE ION, OXYGEN MOLECULE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Tuning Enzyme Thermostability via Computationally Guided Covalent Stapling and Structural Basis of Enhanced Stabilization.
Biochemistry, 61, 2022
|
|
8G3H
 
 | |
7URG
 
 | |
5V1Q
 
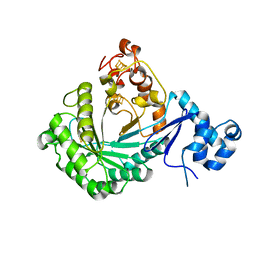 | | Crystal structure of Streptococcus suis SuiB | | Descriptor: | IRON/SULFUR CLUSTER, Radical SAM | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V1S
 
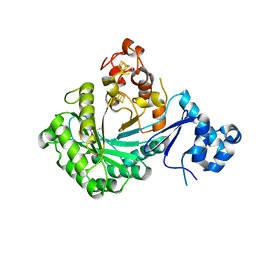 | |
5V1T
 
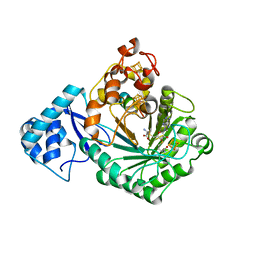 | | Crystal structure of Streptococcus suis SuiB bound to precursor peptide SuiA | | Descriptor: | IRON/SULFUR CLUSTER, METHIONINE, Radical SAM, ... | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6MV9
 
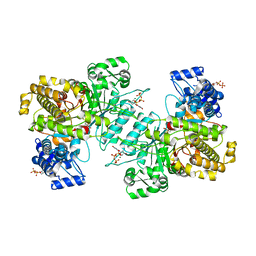 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MT9
 
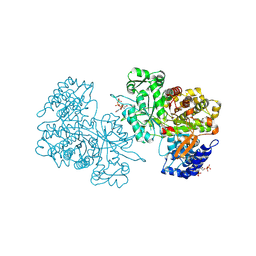 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MVE
 
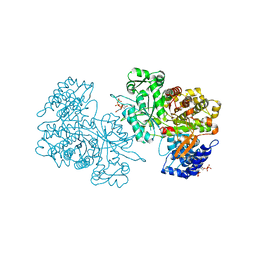 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MYX
 
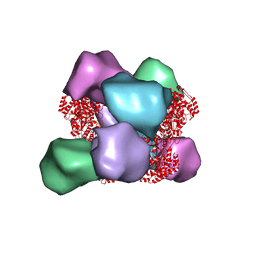 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6O2H
 
 | |
6MW3
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
