6L11
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
8HE4
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HTF
 
 | | Crystal structure of an effector in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTD
 
 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTC
 
 | | Crystal structure of a SeMet-labeled effector from Chromobacterium violaceum in complex with Ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8J8P
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8J8Q
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
5AX8
 
 | | Recombinant expression, purification and preliminary crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial | | Authors: | Jiang, X, Wang, J, Chang, H, Zhou, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase
Biosci Trends, 10, 2016
|
|
6IGY
 
 | |
6JAW
 
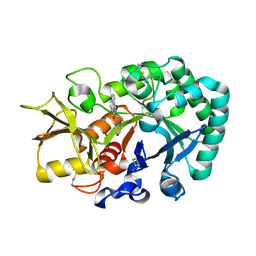 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a napthalimide derivative | | Descriptor: | 2-[3-(morpholin-4-yl)propyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAV
 
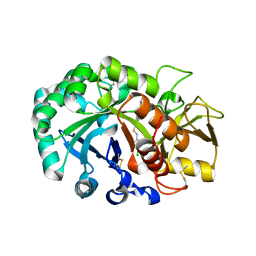 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAY
 
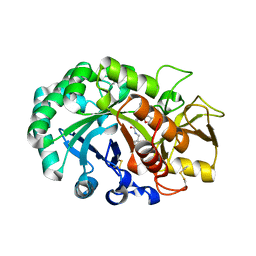 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a dipyrido-pyrimidine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-[(furan-2-yl)methyl]-5-oxo-3-({[(2S)-oxolan-2-yl]methyl}carbamoyl)-5H-dipyrido[1,2-a:2',3'-d]pyrimidin-1-ium, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAX
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6NJD
 
 | |
6NII
 
 | |
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
