3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | Descriptor: | Alkaline serine protease ver112 | | Authors: | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-09 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
4F7G
 
 | |
4YZ9
 
 | |
4YZC
 
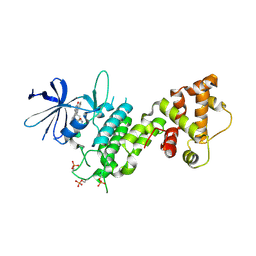 | | Crystal structure of pIRE1alpha in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
4YZD
 
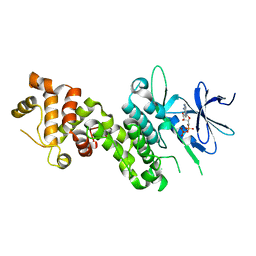 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
7F7W
 
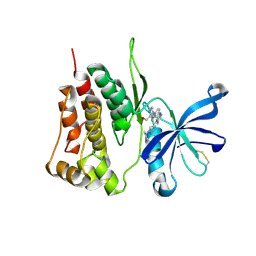 | | JAK2-JH2 | | Descriptor: | 2-((1-(2-fluoro-4-((4-(1-isopropyl-1H-pyrazol-4-yl)-5-methylpyrimidin-2-yl)amino)phenyl)piperidin-4-yl)(methyl)amino)ethan-1-ol, Tyrosine-protein kinase JAK2 | | Authors: | Niu, L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preclinical studies of Flonoltinib Maleate, a novel JAK2/FLT3 inhibitor, in treatment of JAK2 V617F -induced myeloproliferative neoplasms.
Blood Cancer J, 12, 2022
|
|
7EXC
 
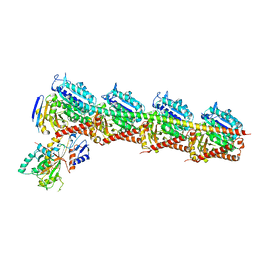 | | Crystal structure of T2R-TTL-1129A2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Design and Synthesis of N-Substituted 3-Amino-beta-Carboline Derivatives as Potent alpha beta-Tubulin Degradation Agents
J.Med.Chem., 65, 2022
|
|
7CBZ
 
 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6JHY
 
 | |
5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
4GMB
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-402 | | Descriptor: | MM-402, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J.Med.Chem., 60, 2017
|
|
7BVP
 
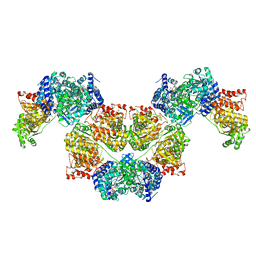 | | AdhE spirosome in extended conformation | | Descriptor: | Aldehyde-alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Kim, G.J, Song, J.J. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling.
Commun Biol, 3, 2020
|
|
7CNC
 
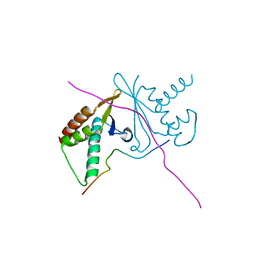 | | cystal structure of human ERH in complex with DGCR8 | | Descriptor: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | Authors: | Li, F, Shen, S. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
7CI1
 
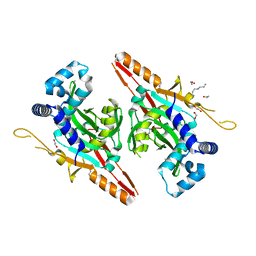 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
7CI2
 
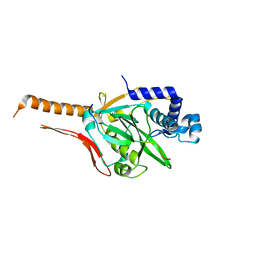 | |
5ULA
 
 | |
8GYA
 
 | |
8GY9
 
 | |
8GYB
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
3KGK
 
 | | Crystal structure of ArsD | | Descriptor: | Arsenical resistance operon trans-acting repressor arsD | | Authors: | Ye, J, Rosen, B.P. | | Deposit date: | 2009-10-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase.
Biochemistry, 49, 2010
|
|
7U8Z
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
6R48
 
 | |
