4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | Descriptor: | IODIDE ION, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
2PQJ
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32), complex with adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Au, S.W.N, Cha, S.S, Young, J.A, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2W1O
 
 | | NMR structure of dimerization domain of human ribosomal protein P2 | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Chan, D.S, Sze, K.H, Zhu, G, Shaw, P.C, Wong, K.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Dimerization Domain of Ribosomal Protein P2 Provides Insights for the Structural Organization of Eukaryotic Stalk.
Nucleic Acids Res., 38, 2010
|
|
2WGL
 
 | |
4HI0
 
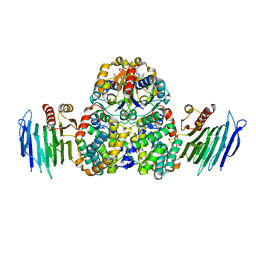 | | Crystal Structure of Helicobacter pylori Urease Accessory Protein UreF/H/G complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Urease accessory protein UreF, Urease accessory protein UreG, ... | | Authors: | Fong, Y.H, Chen, Y.W, Wong, K.B. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease.
Plos Biol., 11, 2013
|
|
1W41
 
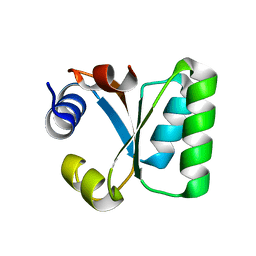 | | T. celer L30e E90A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1W40
 
 | | T. celer L30e K9A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1W42
 
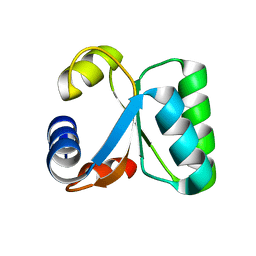 | | T. celer L30e R92A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1NLI
 
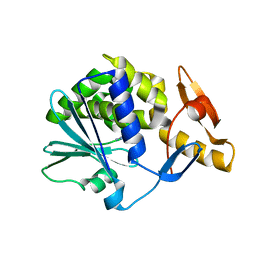 | | Complex of [E160A-E189A] trichosanthin and adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein alpha-trichosanthin | | Authors: | Shaw, P.C, Wong, K.B, Chan, D.S.B, Williams, R.L. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for the interaction of [E160A-E189A]-trichosanthin with adenine.
Toxicon, 41, 2003
|
|
3SF5
 
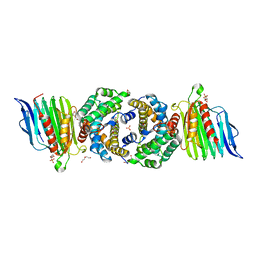 | | Crystal Structure of Helicobacter pylori Urease Accessory Protein UreF/H complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Fong, Y.H, Chen, Y.W, Wong, K.B. | | Deposit date: | 2011-06-12 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF-UreH protein complex
J.Biol.Chem., 286, 2011
|
|
7VB2
 
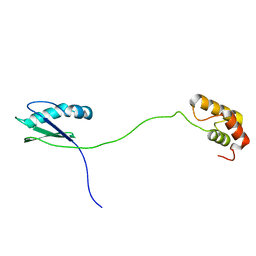 | | Solution structure of human ribosomal protein uL11 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Lee, K.M, Wong, K.B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The flexible N-terminal motif of uL11 unique to eukaryotic ribosomes interacts with P-complex and facilitates protein translation.
Nucleic Acids Res., 50, 2022
|
|
4BEH
 
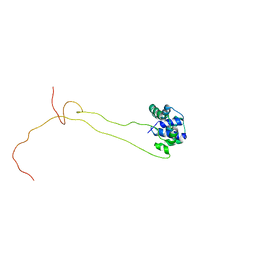 | | Solution structure of human ribosomal protein P1.P2 heterodimer | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P1, 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Yusa, K, Chu, L.O, Wing-Heng Yu, C, Shaw, P.C, Oono, M, Miyoshi, T, Ito, K, Wong, K.B, Uchiumi, T. | | Deposit date: | 2013-03-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human P1P2 Heterodimer Provides Insights Into the Role of Eukaryotic Stalk in Recruiting the Ribosome-Inactivating Protein Trichosanthin to the Ribosome.
Nucleic Acids Res., 41, 2013
|
|
7XRW
 
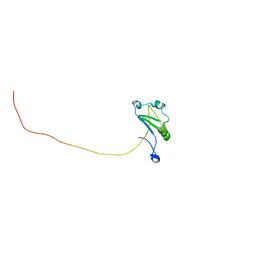 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
3O1Q
 
 | |
5GU4
 
 | | rRNA N-glycosylase RTA | | Descriptor: | GLY-PHE-GLY-LEU-PHE-ASP, GLYCEROL, Ricin | | Authors: | Shi, W.W, Tang, Y.S, Sze, S.Y, Zhu, Z.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2016-08-24 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Ribosome-Inactivating Protein Ricin A Chain in Complex with the C-Terminal Peptide of the Ribosomal Stalk Protein P2
Toxins, 8, 2016
|
|
2JG7
 
 | | Crystal structure of Seabream Antiquitin and Elucidation of its substrate specificity | | Descriptor: | ANTIQUITIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tang, W.K, Wong, K.B, Cha, S.S, Lee, H.S, Cheng, C.H.K, Fong, W.P. | | Deposit date: | 2007-02-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Seabream Antiquitin Reveals the Structural Basis of its Substrate Specificity.
FEBS Lett., 582, 2008
|
|
3RA6
 
 | |
3TLO
 
 | |
3VB7
 
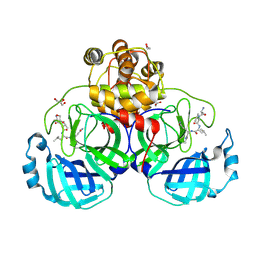 | | Crystal structure of SARS-CoV 3C-like protease with M4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, GLYCEROL, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB6
 
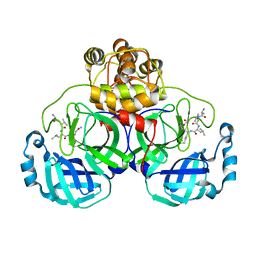 | | Crystal structure of SARS-CoV 3C-like protease with C6Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C6Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB3
 
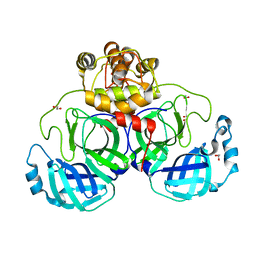 | | Crystal structure of SARS-CoV 3C-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, DI(HYDROXYETHYL)ETHER | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB5
 
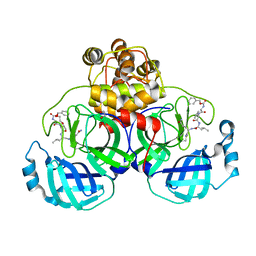 | | Crystal structure of SARS-CoV 3C-like protease with C4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C4Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB4
 
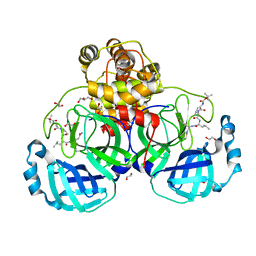 | | Crystal structure of SARS-CoV 3C-like protease with B4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, B4Z inhibitor, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3N4Z
 
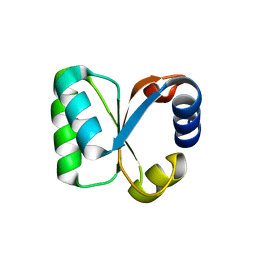 | |
3N4Y
 
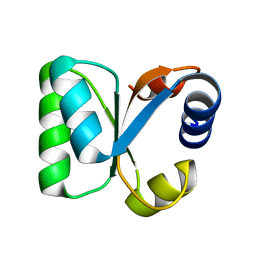 | |
