8FHC
 
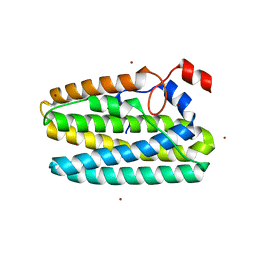 | | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium | | Descriptor: | BROMIDE ION, CHOLIC ACID, FE (III) ION, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium
To Be Published
|
|
3OM9
 
 | |
4YL2
 
 | | Aerococcus viridans L-lactate oxidase Y191F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-03-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Speeding up the product release: a second-sphere contribution from Tyr191 to the reactivity of l-lactate oxidase revealed in crystallographic and kinetic studies of site-directed variants.
Febs J., 282, 2015
|
|
4RJE
 
 | | Aerococcus viridans L-lactate oxidase mutant | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Lactate oxidase, ... | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Ala95-to-Gly substitution in Aerococcus viridans l-lactate oxidase revisited - structural consequences at the catalytic site and effect on reactivity with O2 and other electron acceptors.
Febs J., 282, 2015
|
|
6D9F
 
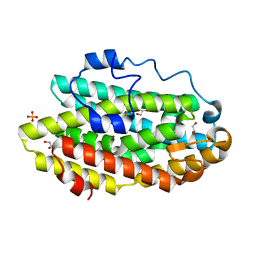 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
3C2E
 
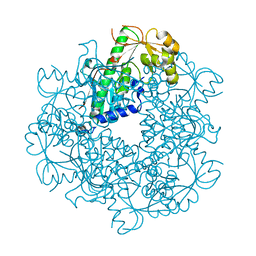 | |
3C2R
 
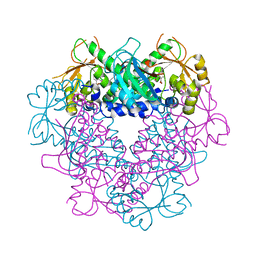 | |
3C2O
 
 | |
3C2F
 
 | |
3C2V
 
 | |
5VM6
 
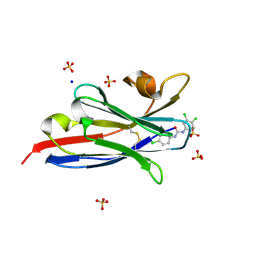 | | The hapten triclocarban bound to the single domain camelid nanobody VHH T10 | | Descriptor: | N-(4-chlorophenyl)-N'-(3,4-dichlorophenyl)urea, SODIUM ION, SULFATE ION, ... | | Authors: | Tabares-da Rosa, S, Gonzalez-Sapienza, G, Wilson, D.K. | | Deposit date: | 2017-04-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of several triclocarban-binding single domain camelid antibody fragments.
J. Mol. Recognit., 32, 2019
|
|
5VL2
 
 | |
5VM4
 
 | |
5VM0
 
 | | The hapten triclocarban bound to the single domain camelid nanobody VHH T9 | | Descriptor: | 1,2-ETHANEDIOL, Camelid Nanobody VHH T9, N-(4-chlorophenyl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Wogulis, L.A, Wogulis, M.D, Tabares-da Rosa, S, Gonzalez-Sapienza, G, Wilson, D.K. | | Deposit date: | 2017-04-26 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and specificity of several triclocarban-binding single domain camelid antibody fragments.
J. Mol. Recognit., 32, 2019
|
|
5VLV
 
 | |
4INW
 
 | |
5ABP
 
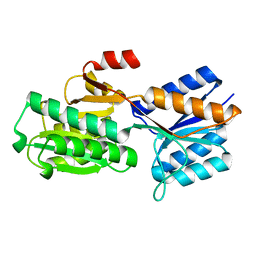 | |
3WCZ
 
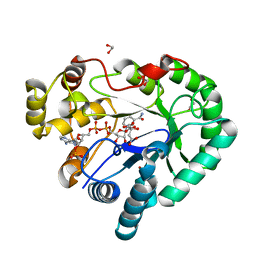 | | Crystal structure of Bombyx mori aldo-keto reductase (AKR2E4) in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase 2E, CITRIC ACID, ... | | Authors: | Yamamoto, K, Wilson, D.K. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification, characterization, and crystal structure of an aldo-keto reductase (AKR2E4) from the silkworm Bombyx mori.
Arch.Biochem.Biophys., 538, 2013
|
|
2ERB
 
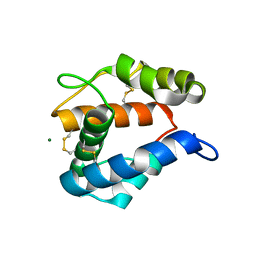 | | AgamOBP1, and odorant binding protein from Anopheles gambiae complexed with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, MAGNESIUM ION, odorant binding protein | | Authors: | Wogulis, M, Morgan, T, Ishida, Y, Leal, W.S, Wilson, D.K. | | Deposit date: | 2005-10-24 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism.
Biochem.Biophys.Res.Commun., 339, 2006
|
|
2FJ0
 
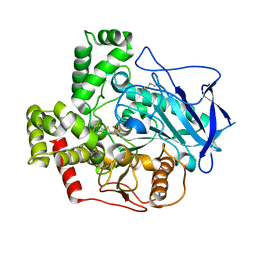 | |
5DIC
 
 | |
5EBU
 
 | | Aerococcus viridans L-lactate oxidase Y215F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-10-19 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational flexibility related to enzyme activity: evidence for a dynamic active-site gatekeeper function of Tyr(215) in Aerococcus viridans lactate oxidase.
Sci Rep, 6, 2016
|
|
2ITM
 
 | | Crystal structure of the E. coli xylulose kinase complexed with xylulose | | Descriptor: | AMMONIUM ION, D-XYLULOSE, SULFATE ION, ... | | Authors: | di Luccio, E, Voegtli, J, Wilson, D.K. | | Deposit date: | 2006-10-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli.
J.Mol.Biol., 365, 2007
|
|
1YE4
 
 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
FEBS Lett., 579, 2005
|
|
1YE6
 
 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NADP+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
Febs Lett., 579, 2005
|
|
