1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
1C1G
 
 | |
1AK5
 
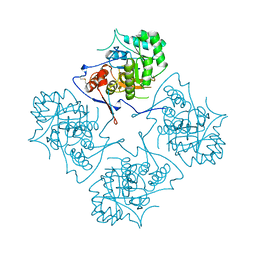 | |
1LC3
 
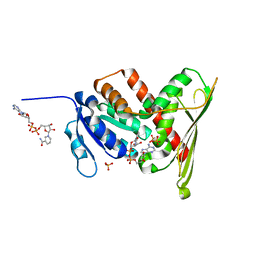 | | Crystal Structure of a Biliverdin Reductase Enzyme-Cofactor Complex | | Descriptor: | Biliverdin Reductase A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Whitby, F.G, Phillips, J.D, Hill, C.P, McCoubrey, W, Maines, M.D. | | Deposit date: | 2002-04-05 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex.
J.Mol.Biol., 319, 2002
|
|
1LC0
 
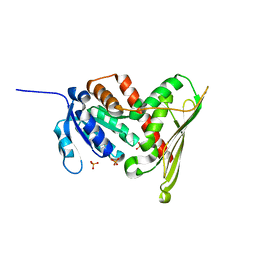 | | Structure of Biliverdin Reductase and the Enzyme-NADH Complex | | Descriptor: | Biliverdin Reductase A, PHOSPHATE ION | | Authors: | Whitby, F.G, Phillips, J.D, Hill, C.P, McCoubrey, W, Maines, M.D. | | Deposit date: | 2002-04-04 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex.
J.Mol.Biol., 319, 2002
|
|
1NDD
 
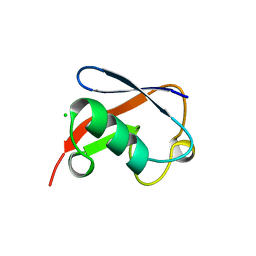 | | STRUCTURE OF NEDD8 | | Descriptor: | CHLORIDE ION, PROTEIN (UBIQUITIN-LIKE PROTEIN NEDD8), SULFATE ION | | Authors: | Whitby, F.G, Xia, G, Pickart, C.M, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes.
J.Biol.Chem., 273, 1998
|
|
1URO
 
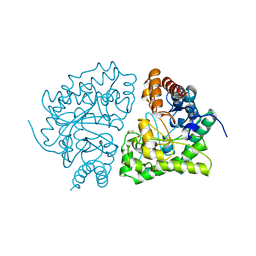 | | UROPORPHYRINOGEN DECARBOXYLASE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (UROPORPHYRINOGEN DECARBOXYLASE) | | Authors: | Whitby, F.G, Phillips, J.D, Kushner, J.P, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human uroporphyrinogen decarboxylase.
EMBO J., 17, 1998
|
|
7RCM
 
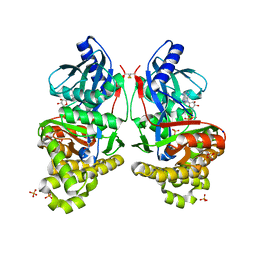 | | Crystal Structure of ADP-bound Galactokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Galactokinase, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7RCL
 
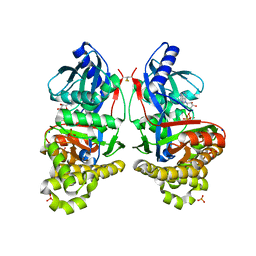 | | Crystal Structure of ADP-bound Galactokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Galactokinase, MAGNESIUM ION, ... | | Authors: | Whitby, F.G, Hall, M.D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7S4C
 
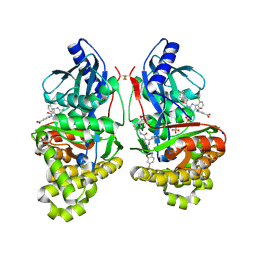 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | 2-({(4R)-4-(2-chlorophenyl)-2-[(6-fluoro-1,3-benzoxazol-2-yl)amino]-6-methyl-1,4-dihydropyrimidine-5-carbonyl}amino)pyridine-4-carboxylic acid, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7S49
 
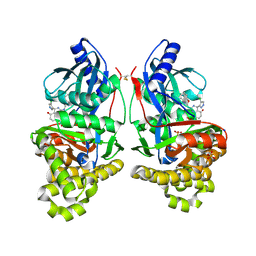 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | (4R)-2-[(1,3-benzoxazol-2-yl)amino]-4-(4-chloro-1H-pyrazol-5-yl)-4,6,7,8-tetrahydroquinazolin-5(1H)-one, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5ILS
 
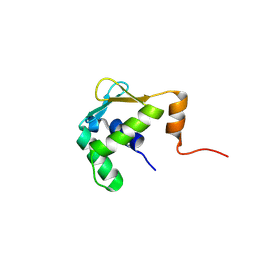 | | Autoinhibited ETV1 | | Descriptor: | ETS translocation variant 1 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILV
 
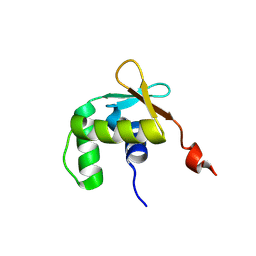 | | Uninhibited ETV5 | | Descriptor: | ETS translocation variant 5 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILU
 
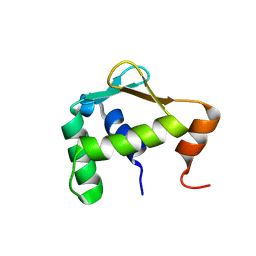 | | Autoinhibited ETV4 | | Descriptor: | ETS translocation variant 4 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
7SMU
 
 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
6E93
 
 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
8UC6
 
 | | Calpain-7:IST1 Complex | | Descriptor: | Calpain-7, IST1 homolog | | Authors: | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
7TXE
 
 | | Plasmodium falciparum Cyt c2 DSD | | Descriptor: | Cytochrome c2, HEME C | | Authors: | Hill, C.P, Wienkers, H.J, Whitby, F.G. | | Deposit date: | 2022-02-08 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites
Proc Natl Acad Sci U S A, 120, 2023
|
|
7U2V
 
 | |
6E94
 
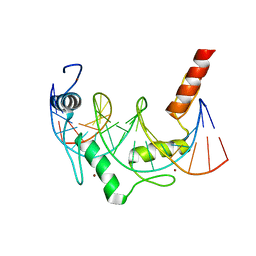 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 K1055R in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
2HTH
 
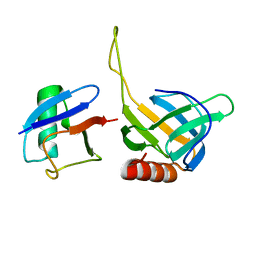 | | Structural basis for ubiquitin recognition by the human EAP45/ESCRT-II GLUE domain | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Alam, S.L, Whitby, F.G, Hill, C.P, Sundquist, W.I. | | Deposit date: | 2006-07-25 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3C3O
 
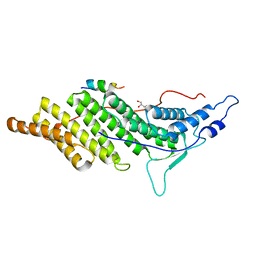 | | ALIX Bro1-domain:CHMIP4A co-crystal structure | | Descriptor: | Charged multivesicular body protein 4a peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C3R
 
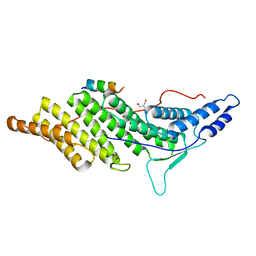 | | ALIX BRO1 CHMP4C complex | | Descriptor: | Charged multivesicular body protein 4c peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | Descriptor: | CHLORIDE ION, Vps4 AAA ATPase | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
