7JUG
 
 | |
7L66
 
 | |
7L62
 
 | |
7L63
 
 | |
7L64
 
 | |
7L65
 
 | |
7L68
 
 | |
7L61
 
 | |
7L67
 
 | |
3OUX
 
 | | Structure of beta-catenin with phosphorylated Lef-1 | | Descriptor: | Catenin beta-1, Lymphoid enhancer-binding factor 1 | | Authors: | Weis, W.I, Sun, J. | | Deposit date: | 2010-09-15 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
J.Mol.Biol., 405, 2011
|
|
3OUW
 
 | | Structure of beta-catenin with Lef-1 | | Descriptor: | Catenin beta-1, Lymphoid enhancer-binding factor 1 | | Authors: | Weis, W.I, Sun, J. | | Deposit date: | 2010-09-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
J.Mol.Biol., 405, 2011
|
|
1YTT
 
 | | YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K | | Descriptor: | MANNOSE-BINDING PROTEIN A, YTTERBIUM (III) ION | | Authors: | Burling, F.T, Weis, W.I, Flaherty, K.M, Brunger, A.T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-06-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of protein solvation and discrete disorder with experimental crystallographic phases.
Science, 271, 1996
|
|
1D2N
 
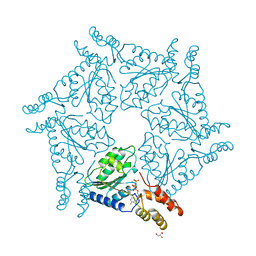 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1DOV
 
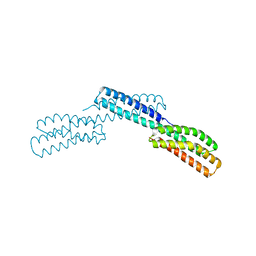 | |
4U15
 
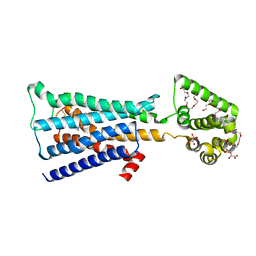 | | M3-mT4L receptor bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, D(-)-TARTARIC ACID, ... | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
4U14
 
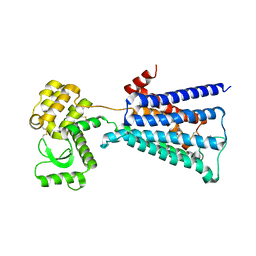 | | Structure of the M3 muscarinic acetylcholine receptor bound to the antagonist tiotropium crystallized with disulfide-stabilized T4 lysozyme (dsT4L) | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3,Endolysin,Muscarinic acetylcholine receptor M3 | | Authors: | Thorsen, T.S, Matt, R.A, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
4U16
 
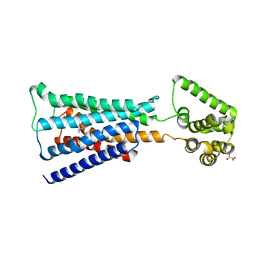 | | M3-mT4L receptor bound to NMS | | Descriptor: | D(-)-TARTARIC ACID, Muscarinic acetylcholine receptor M3,Lysozyme,Muscarinic acetylcholine receptor M3, N-methyl scopolamine | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
1RDN
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-N-ACETYLGLUCOSAMINIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDJ
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH BETA-METHYL-L-FUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDK
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH D-GALACTOSE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDM
 
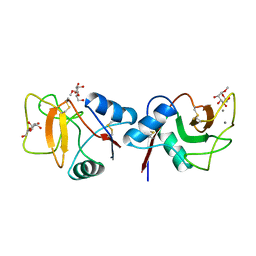 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (1.3 M) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDL
 
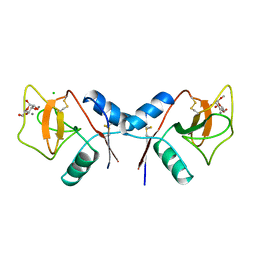 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (0.2 M) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDO
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDI
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-L-FUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1V18
 
 | |
