2XY7
 
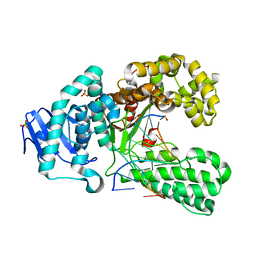 | | Crystal structure of a salicylic aldehyde base in the pre-insertion site of fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP)-3', 5'-D(*SAYP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP)-3', DNA POLYMERASE I, ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY5
 
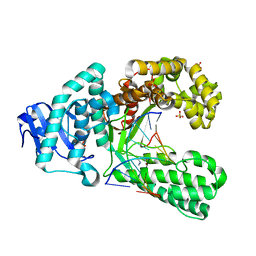 | | Crystal structure of an artificial salen-copper basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*SAYP*TP*CP*CP*CP*TP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2BQW
 
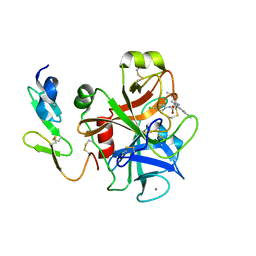 | | CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 | | Descriptor: | 1-{2-[(4-CHLOROPHENYL)AMINO]-2-OXOETHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-28 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BOH
 
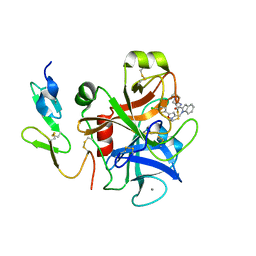 | | Crystal structure of factor Xa in complex with compound "1" | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR XA | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-11 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BQ7
 
 | | Crystal structure of factor Xa in complex with 43 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, FACTOR XA, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BQ6
 
 | | Crystal structure of factor Xa in complex with 21 | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-3-CYANO-N-(1-ISOPROPYLPIPERIDIN-4-YL)-7-METHYL-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
4A7I
 
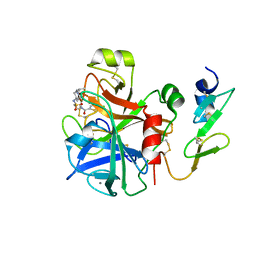 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2XY6
 
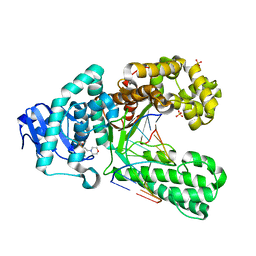 | | Crystal structure of a salicylic aldehyde basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
3NN4
 
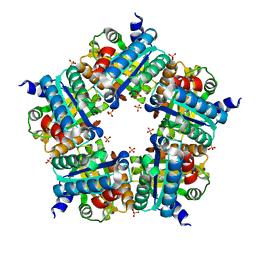 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173K mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN2
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with cyanide | | Descriptor: | CYANIDE ION, Chlorite dismutase, GLYCEROL, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN1
 
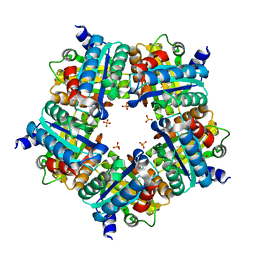 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with imidazole | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN3
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173A mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3QPI
 
 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
9FEK
 
 | |
7NTN
 
 | | The structure of RRM domain of human TRMT2A at 2 A resolution | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Davydova, E, Janowski, R, Witzenberger, M, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
7NTO
 
 | | The structure of RRM domain of human TRMT2A at 1.23 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Witzenberger, M, Janowski, R, Davydova, E, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
5V7R
 
 | |
5V7U
 
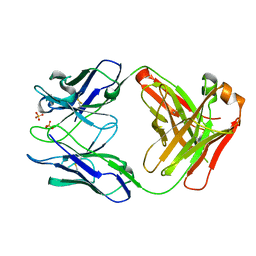 | |
6YAU
 
 | | CRYSTAL STRUCTURE OF ASGPR 1 IN COMPLEX WITH GN-A. | | Descriptor: | 5-[(2~{R},3~{R},4~{R},5~{R},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-~{N}-[3-(propanoylamino)propyl]pentanamide, Asialoglycoprotein receptor 1, CALCIUM ION | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
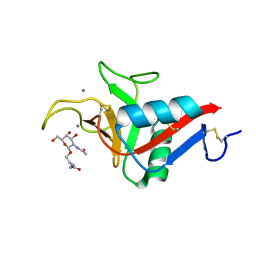
6YG9
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN (HSA) IN COMPLEX WITH GN-07. | | Descriptor: | 20-[[(2~{S})-5-[2-[2-[2-[2-[2-[2-(diethylamino)-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]amino]-20-oxidanylidene-icosanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
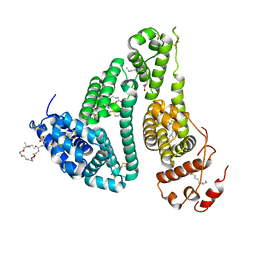
6YSC
 
 | | GLYCOSYLATED KNOB-HOLE/DUMMY FC FRAGMENT | | Descriptor: | Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Leibrock, L, Benz, J. | | Deposit date: | 2020-04-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices.
Nat Commun, 11, 2020
|
|
